Abstract
Celosia cristata is an important economic plant with ornamental, ecological, and medicinal functions. The complete chloroplast genome of C. cristata ‘Xiaguang’ was sequenced and analyzed in this study using Illumina and PacBio sequencing. The chloroplast genome is 153,472 bp in length with a quadripartite structure containing a pair of inverted repeat regions of 25,408 bp (IRa and IRb) separated by a large single-copy region (LSC) of 84,975 bp and a small single-copy region (SSC) of 17,681 bp. The C. cristata ‘Xiaguang’ chloroplast genome encodes 117 genes, comprising 73 protein-coding genes, 36 tRNA genes, and eight rRNA genes. The A + T content of the whole chloroplast genome is 63.3%, whereas the LSC, SSC, and IR region are 65.5%, 69.4%, and 57.4%, respectively.
Celosia cristata belongs to the genus Celosia in the family Amaranthaceae. C. cristata ‘Xiaguang’ was planted in the Environmental Horticulture Research Institute of the Guangdong Academy of Agricultural Sciences (N23°23′, E113°23′, Guangzhou, China). ‘Xiaguang’ cockscomb is a new cultivar developed from the seeds of a recoverable transposon line of C. cristata ‘Zaoshui’, a traditional cultivar in Guangdong (N23°06′, E113°14′). ‘Zaoshui’ was stored in the Environmental Horticulture Research Institute, Guangdong Academy of Agricultural Sciences. On 30 October 2013, experts from the Guangdong Provincial General Station of Seed Management organized the on-site identification of the variety ‘Xiaguang Cockscomb’ bred by the Environmental Horticulture Research Institute of the Guangdong Academy of Agricultural Sciences, and it was approved by the Guangdong Provincial Crop Variety Approval Committee in January 2014 (Fixed. No. 2014006).
We first extracted the chloroplast genome DNA from young leaves of C. cristata ‘Xiaguang’, and a Covaris M220 (Covaris, Woburn, MA, USA) was used to break the DNA into 300–500 bp fragments. Second, shotgun sequencing libraries were constructed according to the TruSeq™ DNA Sample Prep Kit for Illumina. Third, whole genome sequencing was executed using the Illumina HiSeq 4000 platform. The Illumina sequencing data were preliminarily assembled using SOAPdenovo (v2.04) (Luo et al. Citation2012), and subsequent analysis was carried out using Celera Assembler (version 8.0, Rockville, MD, USA). The complete cp genome sequence has been submitted to Genbank with the accession number of MK470118.
The complete chloroplast genome sequence of C. cristata ‘Xiaguang’ is 153,472 bp in length, including two inverted repeat regions (IRa and IRb, each 25,408 bp) separated by an SSC (17,681 bp) region and an LSC (84,975 bp) region. The GC content of the overall chloroplast genome, IR regions, SSC, and LSC are 36.73, 34.47, 30.61, and 42.64%, respectively. The GC content of the two IR regions is higher than those of the SSC and LSC, which is very common in other plants; this phenomenon is mostly attributable to tRNA and rRNA genes (Mardis Citation2013; Asaf et al. Citation2017). The chloroplast genome contains 117 genes in total, including 73 protein-coding genes, 36 tRNAs, and eight rRNAs. The IR region includes six protein-coding genes, seven tRNAs, and four rRNAs. The SSC contains only one tRNA and 10 protein-coding genes, whereas the LSC contains 57 protein-coding genes and 21 tRNAs.
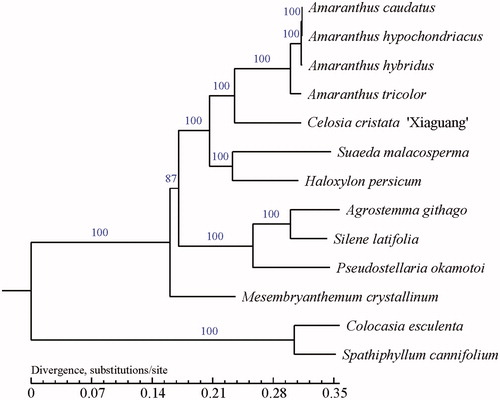
An evolutionary tree was constructed based on the population SNP matrix of the sample and reference genomes. PhyML v3.0 (http://www.atgc-montpellier.fr/phyml/) and 1000 bootstrap replicates were used to construct a phylogenetic tree using the maximum-likelihood (ML) method (). This study will be useful for further analysis on genetic diversity, molecular markers, and molecular breeding.
Figure 1. ML phylogenetic tree reconstruction of 13 species based on sequences from whole chloroplast genomes. GenBank accession numbers for each plant species are as follows: C. Cristata ‘Xiaguang’ (MK470118), Amaranthus tricolor (KX094399), Amaranthus hypochondriacus (MG836505), Amaranthus hybridus (MG836507), Amaranthus caudatus (MG836508), Haloxylon persicum (NC_027669), Mesembryanthemum (NC_029049), Agrostemma githago (KF527884), Silene latifolia (KT962040), Pseudostellaria okamotoi (MH879018), Suaeda malacosperma (NC_039180), Colocasia esculenta (NC_016753), and Spathiphyllum cannifolium (MK372232), with the last two species used as outgroups.
Author contributions
Performed the experiments investigation, project administration, writing the original draft and data curation: XL.
Prepared the resources: YY, JL, and BY.
Supervised the project and made revisions to the manuscript: YX.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Asaf S, Khan AL, Khan MA, Waqas M, Kang SM, Yun BW, Lee IJ. 2017. Chloroplast genomes of Arabidopsis halleri ssp. gemmifera and Arabidopsis lyrata ssp. petraea: structures and comparative analysis. Sci Rep. 7(1):7556.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18–24.
- Mardis ER. 2013. Next-generation sequencing platforms. Ann Rev Anal Chem. 6(1):287–303.
