Abstract
The complete plastid genome of Gentiana filistyla, occurring in Hengduan Mountains with high altitudes, was determined and analyzed in this work. It had a circular-mapping molecular with the length of 139,476 bp, has similar gene composition with G. section Kudoa but contains eight less ndh genes than the available species from sections Cruciata, Microsperma and Frigidae. Phylogenetic analysis showed that G. filistyla clustered together with section Kudoa rather than sections Microsperma and Frigidae. The plastome provided in this work would be useful for elucidation of phylogenetics and evolution in Gentiana.
As a big genus containing 362 species (Ho and Liu Citation2001), Gentiana plants are typically alpine and important parts of alpine ecosystem. Gentiana filistyla I. B. Balfour and Forrest, belonging to section Isomeria Kusnezow series Uniflorae Marquand, is endemic to Hengduan Mountains with very narrow distribution (Ho and Liu Citation2001). Currently, there has been no genomic studies in section Isomeria.
Herein, we reported and characterized the complete platome of G. filistyla (MN199134). One G. filistyla individual (specimen number: Fu2018093-3) was collected from Chayu, Tibet, China (28°36′N, 98°04′E, 4640 m/a.s.l) and its voucher specimens was deposited in the herbarium of Luoyang Normal University. The fragmented genomic DNA was sequenced using Illumina HiSeq 2500 platform (Novogene, Tianjing, China), yielding approximately 3 Gb of 150-bp paired-end. The plastome was de novo assembled in NOVOPlasty 2.6.1 (Dierckxsens et al. Citation2017) and then annotated in GeSeq (Tillich et al. Citation2017) using the default parameters. Comparative analysis was conducted in mVISTA (Frazer et al. Citation2004) with species from four plastome-available Gentiana sections, Kudoa (Sun et al. Citation2018), Cruciata (Zhou et al. Citation2018), Microsperma (Sun et al. Citation2019b) and Frigidae (Sun et al. Citation2019a). Shared protein coding genes in plastomes of available Gentiana species were extracted and aligned using MAFFT (Katoh et al. Citation2002). Using concatenated protein coding genes, maximum likelihood phylogenetic analyses were conducted with IQ-TREE (Nguyen et al. Citation2015) in PhyloSuite (Zhang et al. Citation2020) with 1000 replicates. Swertia mussotii (KC875852) and S. verticillifolia (MF795137) were served as the outgroups.
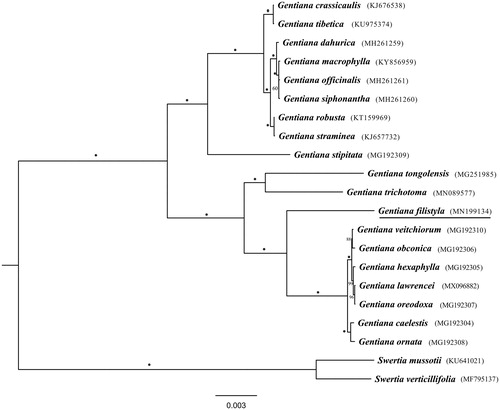
The complete G. filistyla plastome is a circular-mapping molecule with the length of 139,476 bp. The LSC, IR and SSC regions were 77,928, 25,516 and 10,516 bp, respectively. A total of 123 genes were annotated, containing 81 protein-coding genes, 34 tRNA genes and 8 rRNA genes. Comparison analysis indicated that gene composition of G. filistyla platome is similar with section Kudoa (Sun et al. Citation2018), but different with species in section Cruciata (Zhou et al. Citation2018) due to absence of seven ndh genes. The hotspots among sections Isomeria, Kudoa, Cruciata, Microsperma and Frigidae located at intergenic regions such as trnH-GUG–psbA, trnK-UUU–rps16, atpH–atpI, petN–trnD and trnL-UAG–ccsA. Phylogenetic analysis showed that G. filistyla and section Kudoa clustered together into a clade being sister to sections Microsperma and Frigidae (), which is inconsistent with previous study (Favre et al. Citation2016), indicating that more taxa should be involved for further study. The determination of the G. filistyla plastome sequences provided new molecular data to illuminate the phylogenetics and evolution of Gentiana.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Favre A, Michalak I, Chen C-H, Wang J-C, Pringle JS, Matuszak S, Sun H, Yuan Y-M, Struwe L, Muellner-Riehl AN. 2016. Out-of-Tibet: the spatio-temporal evolution of Gentiana (Gentianaceae). J Biogeogr. 43(10):1967–1978. doi:10.1111/jbi.12840.
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32(Web Server):W273–W279.
- Ho TN, Liu SW. 2001. A worldwide monograph of Gentiana. Beijing: Science Press.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Sun SS, Fu PC, Zhou XJ, Cheng YW, Zhang FQ, Chen SL, Gao QB. 2018. The complete plastome sequences of seven species in Gentiana sect. Kudoa (Gentianaceae): insights into plastid gene loss and molecular evolution. Front Plant Sci. 9:493.
- Sun SS, Wang H, Fu PC. 2019a. Complete plastid genome of Gentiana trichotoma (Gentianaceae) and phylogenetic analysis. Mitochondrial DNA B. 4(2):2775–2776.
- Sun SS, Zhou XJ, Li ZZ, Song HY, Long ZC, Fu PC. 2019b. Intra-individual heteroplasmy in the Gentiana tongolensis plastid genome (Gentianaceae). PeerJ. 7:e8025.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Zhang D, Gao F, Li WX, Jakovlić I, Zou H, Zhang J, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355.
- Zhou T, Wang J, Jia Y, Li W, Xu F, Wang X. 2018. Comparative chloroplast genome analyses of species in Gentiana section Cruciata (Gentianaceae) and the development of authentication markers. IJMS. 19(7):1962.