Abstract
We determined the complete mitochondrial genome sequences of an asteroid Linckia laevigata belonging to the order Valvatida. The complete mitogenome of L. laevigata was 16,371 bp in length and consisted of 13 protein-coding genes (PCGs), two rRNA, and 22 tRNA. The orders of PCGs and rRNAs were identical to those of the recorded mitogenomes of asteroids. Phylogenetic analyses placed L. laevigata as the sister group to the species of the other Paxillosida.
Linckia laevigata is a common sea star in the shallow waters of the tropical Indo-West Pacific. Otwoma and Kochzius (Citation2016) conducted phylogeographic studies of the L. laevigata to clarify the connectivity among populations in the Indo-West Pacific. Including such a common, well-studied species, the phylogenetic studies of the superorder Valvatacea are ongoing (e.g. Mah and Foltz Citation2011), and further genetic markers and OTUs are needed for the studies. In non-model organisms like L. laevigata, we have to use universal primers though these primers often fail to amplify a particular region in PCR. To make a specific primer, genomic information of more closely related species is necessary. Usually, mitogenome is including 13 protein-coding genes and two rRNA genes frequently used for phylogenetic reconstruction at every taxonomic level. There was no mitogenome record for the family Ophidiasteridae in Valvatacea, and we choose L. laevigata for the representative of the family. We determined the whole mitogenome sequence and also nucleic rRNA genes by a shotgun sequencing, which is a PCR independent method.
Total DNA was extracted using DNeasy Blood & Tissue Kit (QIAGEN) and processed using the QIAseq FX DNA Library kit (QIAGEN). Paired-end sequencing (300 cycles) was conducted using MiSeq (Illumina) of the National Museum of Nature and Science, with inserts of ca. 50–200 bp for a total of approximate two million reads. Assembly was performed using CLC Genomics Workbench ver. 12 (QIAGEN) with the default setting. Ambiguous part of the contig was confirmed using 3500xL Genetic Analyzer (Thermo Fisher). Gene identification was made using the MITOS web server (Bernt et al. Citation2013). A voucher specimen with extracted DNA was deposited to the National Museum of Nature and Science, Tokyo (NSMT E-12913).
The mitogenome of L. laevigata (GenBank/DDBJ/EMBL accession number LC505032) is 16,371 bp long and encodes 13 proteins, two rRNAs, and 22 tRNAs for a total of 37 gene products. The overall A + T content of the L. laevigata mitochondrial genome is 63.5%, which is around average among asteroidean species (Mu et al. Citation2018). Similar to other sea-star mitogenomes, ND1 and ND2 start with GTG codon, and ND4L starts with ATT codon, and all other protein-coding genes (PCGs) start with the ATG start codon. Eight of PCGs stop with the termination codon TAA, COX3 and ND6 end with TAG codon, whereas other PCGs (COX2, CytB, and ND1) used incomplete termination codon T—.
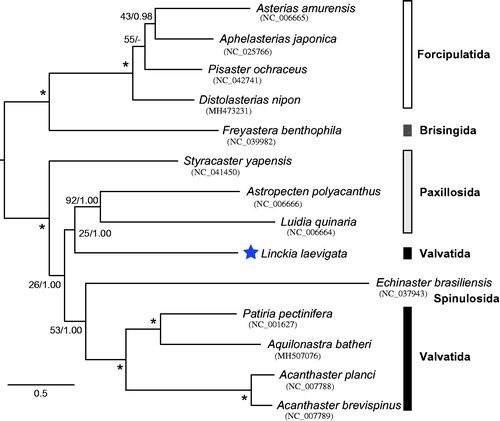
The maximum-likelihood phylogenetic analysis (ML) based on 13 PCGs was conducted using RAxML-NG ver.0.9.0 (Kozlov et al. Citation2019) with bootstrap analyses of 1000 replicates. The phylogenetic tree also with posterior probability from Bayesian analyses (BA) conducted using MrBayes 3.2.6 (Ronquist et al. Citation2012). Linckia laevigata make a sister clade with species belonging to order Paxillosida (Astropecten polyacanthus and Luidia quinaria) with low nodal support value (). The family Ophidasterida, which including L. laevigata, was paraphyletic in the previous study. Although additional OTUs are needed, this mitogenome would be useful for reconstructing higher systematics of asteroid phylogeny.
Figure 1. Maximum-likelihood tree based on the concatenated nucleotide sequence of 13 protein-coding genes of Linckia laevigata (LC505032) and 12 asteroid species. Nodal values are ML bootstrap support values (BS) and BA posterior probabilities (PP). An asterisk (*) indicates 100% BS and 1.0 PP. A hyphen (-) shows the branch not supported in the BA tree. The scale bar indicates branch length in substitutions per site.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Kozlov A, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35(21):4453–4455.
- Mah C, Foltz D. 2011. Molecular phylogeny of the Valvatacea (Asteroidea: Echinodermata). Zool J Linne Soci. 161(4):769–788.
- Mu W, Liu J, Zhang H. 2018. The first complete mitochondrial genome of the Mariana Trench Freyastera benthophila (Asteroidea: Brisingida: Brisingidae) allows insights into the deep-sea adaptive evolution of Brisingida. Ecol Evol. 8(22):10673–10686.
- Otwoma LM, Kochzius M. 2016. Genetic population structure of the coral reef sea star Linckia laevigata in the Western Indian Ocean and Indo-West Pacific. PLoS One. 11(10):e0165552.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
