Abstract
The complete chloroplast genome of Uraria lagopodioides is determined in this study. The plastome is 149,828 bp in length and comprises a large single-copy region (83,133 bp), a small single-copy region (18,443 bp), and a pair of inverted repeats (24,126 bp). A total of 128 genes were identified, including 83 protein-coding genes, 37 tRNA genes, and eight rRNA genes. The overall GC content is 35.2%. Phylogenetic analysis of 15 plastome sequences within Fabaceae revealed U. lagopodioides was closely related to Desmodium heterocarpon.
Uraria Desvaux belongs to Fabaceae and consists of ca. 20 species in tropical Africa, Asia, and Australia (Huang et al. Citation2010). Most of the Uraria species are medicinal plants (Rahman et al. Citation2007; Thien et al. Citation2019). However, no chloroplast genome resource is available for this genus. In this study, we reported the chloroplast genome of Uraria lagopodioides (Linnaeus) Candolle, which is a prostrate or spreading perennial herb characterized by its fox-tail-like inflorescence. This first complete chloroplast genome of Uraria will provide valuable information for evolutionary studies of this genus and the related genera.
The sample was collected from Yuanjiang, Yunnan, China (23°32′53.20″N, 102°10′53.04″E, voucher no. ZXL381-1, deposited at College of Forestry, Southwest Forestry University). Total genomic DNA was extracted from silica-dried leaves using a modified CTAB DNA extraction method (Doyle and Doyle Citation1987). Genome sequencing was performed using Illumina Nova Seq 6000 platform at Annoroad Gene Technology (Beijing, China).
The chloroplast genome reads were assembled with NOVOPlasty (Dierckxsens et al. Citation2017) and modified using Geneious Prime v2020.0.4 (https://www.geneious.com). Then, the initial annotation was performed using PGA (Qu et al. Citation2019) and reconfirmed with Geneious Prime v2020.0.4 based on the annotation of Glycine max (L.) Merr. (NC_007942) and Desmodium heterocarpon (L.) DC. (NC_044113).
The complete chloroplast genome of U. lagopodioides (GenBank accession no. MT040621) was 149,828 bp in length, including a large single-copy region (LSC: 83,133 bp), a small single-copy region (SSC: 18,443 bp), and a pair of inverted repeats (IRs: 24,126 bp). This plastome encodes 128 genes, including 83 protein-coding genes, 37 tRNAs, and eight rRNAs. The overall GC content was 35.2%.
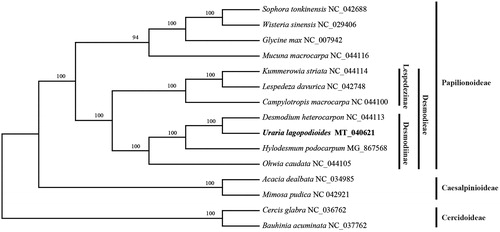
To determine the phylogenetic position of U. lagopodioides within Fabaceae, phylogenetic analysis was conducted along with 14 representative taxa. The 15 chloroplast genome sequences were aligned using MAFFT v7.3 (Kazutaka and Standley Citation2013). Phylogenetic inference using maximum-likelihood (ML) is conducted using GTR + G model with 1000 bootstrap replicates by RAxML v.8.2.10 (Alexandros Citation2014). The results revealed that U. lagopodioides are nested within the Desmodiinae clade which belongs to tribe Desmodieae (Papilionoideae), and it is closely related to Desmodium heterocarpon ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Alexandros S. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang PH, Ohashi H, Iokawa Y. 2010. Flora of China. Vol. 10. Beijing: Science Press & St. Louis: Missouri Botanical Garden Press; p. 286–288.
- Kazutaka K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50.
- Rahman MM, Gibbons S, Gray AI. 2007. Isoflavanones from Uraria picta and their antimicrobial activity. Phytochemistry. 68(12):1692–1697.
- Thien DD, Thuy TT, Anh NTH, Thang LQ, Dai TD, Sa NH, Tam NT. 2019. A new isoflavanone from Uraria crinita. Nat Prod Res. 34:1–7.