Abstract
We here report the complete mitochondrial genome of the Hydrophis melanocephalus slender-necked sea snake from Okinawa, Japan. Its length is 17,182 bp, consisting of 13 protein-coding genes, two rRNA genes, 22 tRNA genes, and three non-coding regions, providing the basis for further phylogenetic and evolutionary studies of sea snakes.
Hydrophis melanocephalus, a rarely studied slender-necked sea snake, has been often confused with other snakes, such as H. coggeri in the Australian region (Kharin Citation1984), Emydocephalus ijimae in Japan (Yi et al. Citation2019), and H. platurus and Laticauda semifasciata in Korea (Kim et al. Citation2018; Kim et al. Citation2020). Hydrophis melanocephalus is known to be distributed in Vietnam, China, Taiwan, Japan, and Philippines (David and Ineich Citation1999; Kharin and Czeblukov Citation2006). Several sightings of the snake have been recorded in Korean coastal regions (Shannon Citation1956; Won Citation1971; Kang and Yoon Citation1975), but no scientific records, such as specimens, photographs, or morphological measurements, exist. In this study, we reported the complete mitochondrial sequence of H. melanocephalus from Okinawa, Japan.
The specimen was captured in Toguchi, Motobu, Kunigami District, Okinawa, Japan (26°39′44.12″ N and 127°53′18.42″ E), and preserved in 70% ethanol at the National Marine Biodiversity Institute of Korea (Voucher No. MABIK AR00000052). Whole genomic DNA was extracted from the muscle tissue using the PCI (Phenol-Chloroform Isoamyl Alcohol) method with DNA extraction solution from Asahida et al. (Citation1996), and proteinase K (Bioneer, Daejeon, South Korea). The whole mitochondrial DNA was sequenced using the Sanger method in a 3730xl DNA analyzer (Applied Biosystems, CA, USA). Sequencing was performed at Macrogen Inc. (Seoul, South Korea). To assemble and annotate the obtained mitochondrial DNA sequences, we used Geneious 9.0.4 (Biomatters Ltd, Auckland, New Zealand), tRNA Scan-SE1.21 software (http://lowelab.ucsc.edu/tRNAScan-SE/) (Lowe and Eddy Citation1997), and Dual Organellar GenoMe Annotator (DOGMA) (Wyman et al. Citation2004). The phylogenetic tree of H. malenocephalus was constructed with PAUP* v4.0b10 (Swofford Citation2003) using the complete mitogenomic sequences, obtained here, as well as from GenBank (). The Bayesian inference and maximum-likelihood method were applied to construct the tree with 1000 bootstrap replicates.
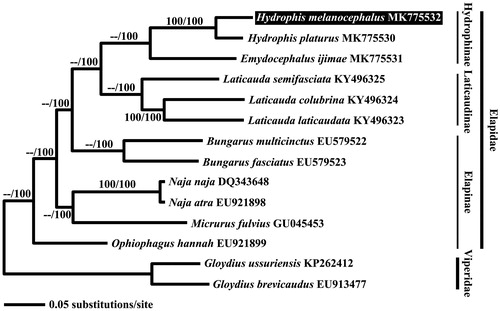
Figure 1. Bayesian inference (BI) tree of Hydrophis melanocephalus based on its 13 mitochondrial protein-coding genes. Both Viperidae species were used as outgroup. The accession number of mitogenomes obtained from GenBank, are indicated after the scientific name of each species. Analyzed values (ML bootstrap value/Bayesian posterior probabilities) are denoted on each branch.
The total length of the H. melanochepalus mitochondrial genome was revealed to be 17,182 bp (GenBank Accession No. MK775532), with a base composition of 33.2% A, 27.3% T, 13.1% G, and 26.4% C, showing an A–T rich (60.5%) feature. The mitogenome contained 13 protein-coding genes (PCGs), two rRNA genes (12S and 16S rRNA), 22 tRNA genes, and three non-coding regions comprised of two control regions, and an L-strand replication origin (OL). Its arrangement pattern and transcribing directions were identical to those of other elapid snakes (Laticauda semifasciata and Emydocephalus ijimae; Kim et al. Citation2018; Yi et al. Citation2019).
The phylogenetic tree revealed that H. melanochepalus formed a sister group with H. platurus, another species of the Hydrophis genus, and showed close relationship with species from the Hydrophinae subfamily, consistent with a previous phylogenetic study of elapid snakes by Lee et al. (Citation2016).
Geolocation information
Toguchi, Motobu, Kunigami District, Okinawa, Japan (26°39′44.12″ N and 127°53′18.42″ E).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction form fish stored at ambient temperature using buffers containing high concentration of urea. Fisheries Sci. 62(5):727–730.
- David P, Ineich I. 1999. Les serpents venimeux du monde: systematique et repartition. Dumerilia. 3:3–499 (in French).
- Kang YS, Yoon IB. 1975. Illustrated encyclopedia of fauna and flora of Korea. Vol. 17. Amphibia-Reptilia. Seoul (South Korea): Samhwa Publisher (in Korean).
- Kharin VE. 1984. Revision of sea snakes of subfamily of Laticaudinae Cope, 1879 sensu lato (Serpentes, Hydrophiidae). Tr ZINAN SSSR. 124:128–139. (in Russian).
- Kharin VE, Czeblukov VP. 2006. A new revision of sea kraits of family Laticaudidae Cope, 1879 (Serpentes: Colubroidea). Russ J Herpetol. 13:227–241.
- Kim IH, Park J, Suk HY, Bae HG, Min MS, Tsai TS, Park D. 2018. Phylogenetic relationships of three representative sea krait species (Genus Laticauda; elapidae; serpentes) based on 13 mitochondrial genes. Mitochondrial DNA A DNA Mapp Seq Anal. 29:772–777.
- Kim JG, Park J, Yi CH, Kim MS, Cho IY, Kim J, Kim IH. 2020. The complete mitochondrial genome of a yellowbellied sea snake (Hydrophis platurus) (Squamata, Elapidae). Mitochondrial DNA B Resour. 5(1):891–892.
- Lee MS, Sanders KL, King B, Palci A. 2016. Diversification rates and phenotypic evolution in venomous snakes (Elapidae). R Soc Open Sci. 3(1):150277.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964.
- Shannon FA. 1956. The reptiles and amphibians of Korea. Herpetologica. 12.1:22–49.
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony, version 4.0 b10. Sunderland (MA): Sinauer Associates.
- Won HG. 1971. Amphibian and reptiles of Chosun. Pyeongyang: Pyeongyang Printing Office (in Korean).
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yi CH, Park J, Sasai T, Kim HS, Kim JG, Kim MS, Cho IY, Kim IH. 2019. Complete mitochondrial genome of the Ijima’s Sea Snake (Emydocephalus ijimae) (Squamata, Elapidae). Mitochondrial DNA B Resour. 4(2):2658–2659.
