Abstract
Lilium davidii var. unicolor (Hoog) cotton, belonging to the family Liliaceae, has essential medicinal and ornamental values. In this study, the whole chloroplast genome of L. davidii var. unicolor was sequenced for the first time. It has the typical quadripartite circle structure, with a genome size of 152,659 bp. Overall, 110 genes were detected, including 78 protein-coding genes, 28 tRNA genes, and 4 rRNA genes. The phylogenetic tree showed that L. davidii var. unicolor was related to L. callosum, L. amabile, L. lancifolium, L. sp., L. tsingtauense, L. hansonii, and L. cernuum.
Lilium davidii var. unicolor, also called Lanzhou lily (Shang et al. Citation2014), one important genus in Liliaceae that is originated in China. The bulb of L. davidii var. unicolor is widely used in the food industry. Moreover, it has medicinal value on heart and lung diseases (Li et al. Citation2014). Most of the studies on L. davidii var. unicolor focused on cultivation and flower chemical component for its ornamental and economic values (Li et al. Citation2014; Shang et al. Citation2016). None researches had put the focus on the chloroplast genome of L. davidii var. unicolor. In the present study, we sequenced the complete chloroplast genome of L. davidii var. unicolor and explore its internal relationships within the family Liliaceae.
Plantlets of L. davidii var. unicolor (voucher number: SD_JBZDZ) was obtained from in vitro Shandandan germplasm preserved at the Conversation and Utilization of Regional Biological Resources, Yan’an, China. L. davidii var. unicolor was obtained in tube seedling form and were originally from Bali Town, Qilihe District, Lanzhou, Gansu, China (N35°09′91.84″, E103°83′51.67″).
Approximately 10 mg fresh young leaf tissue was used for DNA extraction through Plant Chloroplast DNA Column Extraction Kit (BNT120406, Baiao Laibo Technology Co., Ltd. Beijing, China). More than 15 μg DNA was used for the SMRT-bell sequencing (8 kb-10 kb) and high-throughput sequencing library preparation. Initially, the Illumina sequencing data was assembled using SOAPdenovo v2.04 (Luo et al. Citation2012). Then, the sequencing data were subsequently aligned to PacBio RS sequencing data to remove single base and compilation errors using the blasR software (Sergey et al. Citation2012). After that, the corrected data were assembled using Canu v1.5 (https://canu.es/) (Sergey et al. Citation2012). The whole chloroplast genome sequence of L. davidii var. unicolor was deposited to NCBI (https://www.ncbi.nlm.nih.gov/genbank/) database with the accession number of MK954110.
The result showed that the L. davidii var. unicolor chloroplast genome size was 152,659 bp, with typical quadripartite structure: one large single copy (LSC, 82,060 bp), one small single copy (SSC, 17603 bp), and two inverted repeats (IRs, 26,498 bp). The overall GC content was 37.02%. After removing the duplicate genes, 78 protein-coding genes, 28 tRNA genes, and four rRNA genes remained. Most of the genes did not contain the introns, whereas 19 contained one intron, and four genes (clpP, rps12, rps12-D2, and ycf3) contained two introns. The gene rps12 is a trans-spliced gene with its 5′ end located in the LSC region and the duplicated 3′ end in the IR region.
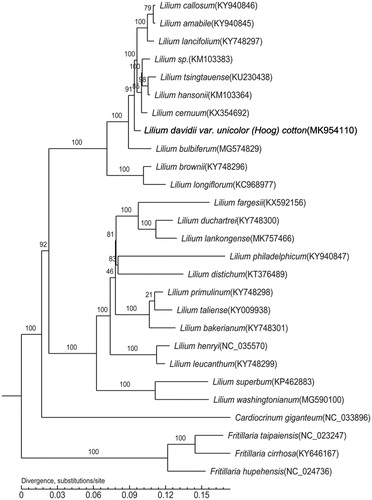
The phylogenetic tree of L. davidii var. unicolor was constructed using the whole chloroplast genome, in combination with other 22 Lilium species and four outgroup species. The phylogenomic relationship was inferred by the Maximum-Likelihood (ML) method based on the general time-reversible (GTR) + G substitution model in PhyML 3.0 (Larkin et al. Citation2007; Guindon et al. Citation2010), with 1000 bootstrap replicates (Letunic and Bork, Citation2016). The phylogenetic tree showed that L. davidii var. unicolor was related to L. callosum, L. amabile, L. lancifolium, L. sp., L. tsingtauense, L. hansonii, and L. cernuum ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Sys Biolo. 59(3):307–321.
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. 2007. Clustal W and clustal X version 2.0. Bioinformatics. 23(21):2947–2948.
- Letunic I, Bork P. 2016. Interactive Tree of Life (ITOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. (web server issue):gkw290.
- Li XY, Wang CX, Cheng JY, Zhang J, Silva J, Liu XY, Duan X, Li TL, Sun HM. 2014. Transcriptome analysis of carbohydrate metabolism during bulblet formation and development in Lilium davidii var. unicolor. BMC Plant Biol. 14(1):358
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18.
- Sergey K, Schatz MC, Walenz BP, Jeffrey M, Howard JT, Ganeshkumar G, Zhong W, Rasko DA, W Richard M, Jarvis ED. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol. 30(7):693–700.
- Shang Q, Yang G, Wang Y, Wu X, Zhao X, Hao H, Li Y, Xie Z, Zhang Y, Wang R. 2016. Illumina-based analysis of the rhizosphere microbial communities associated with healthy and wilted Lanzhou lily (Lilium davidii var. unicolor) plants grown in the field. World J Microbiol Biotecnol. 32:95.
- Shang QH, Zhao X, Li YY, Xie ZK, Wang RY. 2014. First report of Fusarium tircinctum causing stem and root rot on Lanzhou lily (Lilium davidii var. unicolor) in China. Plant Dis. 98(7):999–999.