Abstract
The mitochondrial genome of the epiphytic green alga, Heveochlorella roystonensis was sequenced and characterized. The complete mitogenome contains 130,507 bp, including a single-copy region of 54,865 bp and two direct repeat regions of 37,816 and 37,826 bp, respectively, and has a G + C content of 34.2%. It contains 110 genes, including 73 protein coding genes, 34 tRNA, and three rRNA genes. The large direct repeat makes it currently the largest mitochondrial genome in Trebouxiophyceae.
Heveochlroella roystonensis S. Ma is a terrestrial unicellular green algae identified on the bark surface of the tropical plant royal palm in Haikou, Hainan Province, China with geospatial coordinates N19°59′25″, E110°19′25″ (Ma et al. Citation2013). This species blooms in the wet and warm seasons and plays important roles in the tropical rain forests. The type strain of H. roystonensis is stored at the ClonBank of Institute of Tropical Bioscience and Biotechnology at −80 °C in 15% glycerol under accession number A3-8.
The genomic DNA was extracted using a genomic DNA extraction kit (Tiangen Biotech, Beijing, China), and was sequenced with both PacBio RSII and Illumina Hiseq 2000 platforms. The cleaned PacBio subreads were assembled using CANU (Koren et al. Citation2017), and the possible sequencing errors were corrected using the Illumina Hiseq reads by softwares GATK (Zhu et al. Citation2015). The mitochondrial genome has been deposited in the GenBank under accession number MN934958 and in the Genome Warehouse at the National Genomics Data Center (Members Citation2019), Beijing Institute of Genomics (BIG) under accession number GWHABJT00000000 (https://bigd.big.ac.cn/gwh). The whole genome contains 130,507 bp, including a single-copy region of 54,865 bp and two direct repeat regions of 37,816 and 37,826 bp, and has a G + C content of 34.2%. Compared to the currently known mitogenomes in Trebouxiophyceae, this is the largest genome by containing large direct repeats, which is rare in plants (An et al. Citation2016; Oudot-Le Secq and Green Citation2011). It contains 73 protein-coding genes, including 17 ribosomal protein genes, 11 NAD(P)H-quinone oxidoreductase (nad) genes, two NADH-quinone oxidoreductase (nuoI) genes, seven ATP synthase genes, four cox genes, two cob genes, five intron-encoded endonuclease genes, two lipoprotein signal peptidase genes, and two tatC genes. It contains 34 tRNA genes, in which 12 genes (trnG.1, trnG.2, trnN, trnC, trnY, trnS, trnA, trnP, trnL, trnH, trnW, and trnE) are duplicated due to the direct repeat and the trnG gene has four copies (two in each repeat), while the single copy region contains single copy tRNA genes of trnQ, trnF, trnK, trnI, trnD, trnV, and trnR and two copies of trnM gene, as well as an additional copy of trnL that has been identified in the repeat regions. In total, the 34 tRNA genes correspond to 19 amino acids, but trnT gene is missing. There are three ribosomal RNA genes (rrnL, rrnS, and rrn5). The genes in the two direct repeat regions are located in the same order, however, there are gain or loss of function events between the two repeats. Compared with the mitochondrial genomes of other aquatic Trebouxiophyceae species (), there are more genes present in H. roystonensis corresponding to its adaptability to terrestrial habitats.
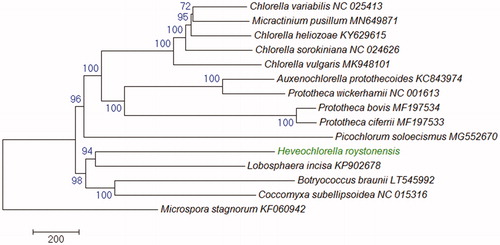
Figure 1. Phylogeneitc relationships of 14 Trebouxiophyceae green algae based on 20 peotein sequences encoded by their mitogenomes. The tree was inferred using the Neighbor-Joining method and drawn with MEGA7 (Kumar et al. Citation2016). Bootstrap supports (1000 replicates) are shown next to the branches. The tree is drawn to scale as indicatd by the scale bar and rooted with a Chlorophyceae species Microspora stagnorum as an outgroup. GenBank accession numbers are shown behind the taxon names.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- An SM, Noh JH, Choi DH, Lee JH, Yang EC. 2016. Repeat region absent in mitochondrial genome of tube-dwelling diatom Berkeleya fennica (Naviculales, Bacillariophyceae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):2137–2138.
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ma S, Huss VAR, Tan D, Sun X, Chen J, Xie Y, Zhang J. 2013. A novel species in the genus Heveochlorella (Trebouxiophyceae, Chlorophyta) witnesses the evolution from an epiphytic into an endophytic lifestyle in tree-dwelling green algae. Eur J Phycol. 48(2):200–209.
- Members BDC. 2019. Database resources of the BIG Data Center in 2019. Nucleic Acids Res. 47(D1):D8–D14.
- Oudot-Le Secq MP, Green BR. 2011. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene. 476(1–2):20–26.
- Zhu P, He L, Li Y, Huang W, Xi F, Lin L, Zhi Q, Zhang W, Tang YT, Geng C, Lu Z, et al. 2015. Correction: OTG-snpcaller: an optimized pipeline based on TMAP and GATK for SNP calling from ion torrent data. PLoS One. 10(9):e0138824.
