Abstract
Ostericum palustre is a critically Endangered species endemic to Europe and was described as the type-species of the genus Ostericum (Apiaceae). The complete chloroplast genome of O. palustre was reported for the first time, which suggested a circle quadripartite structure of 154923 bp in length comprising the large single-copy region (LSC) of 84750 bp, the small single-copy region (SSC) of 19752 bp, and a pair of inverted regions (IRs) of 25209 bp. There were totally 127 genes including 4 rRNAs, 37 tRNAs, and 82 protein-coding genes. In the phylogenetic analyses, O. palustre was very close to Pterygopleurum neurophyllum but distant from Angelica species, which coincided with the result of ITS sequences analysis. This study provided a significant genetic resource and will be helpful to systematic research and effective conservation.
Keywords:
Ostericum palustre Besser was described as the type-species of the genus Ostericum Hoffm. (Apiaceae) which was thought very close to Angelica L. As an endemic herb to Europe, O. palustre was once wide-spread from Russia to Germany and far away from other eight species of Ostericum in East Asia (Shan Citation1992; Sheh et al. Citation2005; Liao et al. Citation2013). Whereas, this umbelliferous plant is seriously threatened by habit loss and declining rapidly during recent decades so that it was listed in the Bern Convention and Annx II of the EU Flora-Fauna Directive-Natura (Schnittler and Gunther Citation1999; Dittbrenner et al. Citation2005; Zych et al. Citation2014). Herein, the complete chloroplast genome of O. palustre was reported in order to offer genomic information for phylogenetical research and effective conservation.
Leaf materials were collected from the cultivated O. palustre (seeds from southern Germany) in greenhouse of Sichuan University, Chengdu, China. The voucher specimens and DNA samples were deposited in the herbarium of Sichuan University under accession no. 15102601(SZ). Morphological characteristics are measured using Karyotype (Altınordu et al. Citation2016). Total genomic DNA was extracted using Plant Genomic DNA Kit. The isolated genomic DNA was manufactured to average 400 bp paired-end (PE) library using the Illumina Hiseq platform (Illumina, San Diego CA, USA), and then sequenced by Illumina genome analyzer (Hiseq PE150). The gene reconstruction and annotation were performed using Geneious Prime 2019.1.1 (Kearse et al. Citation2012), referring to Pterygopleurum neurophyllum (GenBank no. NC033345.1). The complete assemble genome sequence was deposited in GenBank (accession no. MN970215).
The length of the complete chloroplast genome of O. palustre was 154923 bp in a circular form, and comprised four regions: the large single-copy region (LSC) of 84750 bp, the small single-copy region (SSC) of 19752 bp, and two inverted regions (IRs) of 25209 bp each. The total GC content of this species was 38%, with 36% for the LSC, 31% for the SSC and 43% for each IR. There were 4 rRNAs, 37 tRNAs, and 82 protein-coding genes, for a total of 127 genes, contained in the whole chloroplast genome. Among that, 22 tRNA coding genes were placed in the LSC, seven in each IR and only one in the SSC, and the rRNA coding genes exclusively occurred in the IRs.
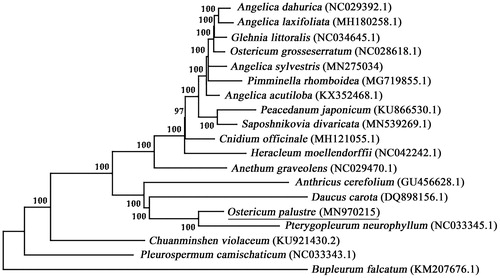
The phylogenetic analysis of O. palustre was conducted with additional 18 accessions of complete chloroplast genome sequences from 15 genera in Apioideae. The dataset were aligned using MAFFT (Katoh et al. Citation2002) and trimmed using trimAl v1.4 (Capella-Gutierrez et al. Citation2009). Then, neighbor-joining (NJ) trees were generated using MEGA7.0 (Kumar et al. Citation2016) with 1000 bootstrap replicates. The topology of the NJ-tree () revealed that O. palustre was located very close to Pterygopleurum neurophyllum but distant from Angelica species, which coincided with previous studies and morphological evidence (Liao et al. Citation2013; Li et al. Citation2017; Zhang et al. Citation2018). However, it was notable that O. grosseserratum (NC028618.1) was distant from O. palustre and allied with Angelica, which was significantly inconsistent with the results of ITS analyses.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Altınordu F, Peruzzi L, Yu Y, He XJ. 2016. A tool for the analysis of chromosomes: KaryoType. Taxon. 65(3):586–592.
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dittbrenner A, Partzsch M, Hensen I. 2005. Beiträgezur Populationsbiologie und Vergesellschaftung von Angelica palustris (Besser) Hoffm.*). Hercynia. 38:59–87.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.27004904
- Li ML, Liao CY, Ye C, Zhang JZ, Feng TY, Zhou B. 2017. Analysis of leaf epidermal micromorphological characters of Ostericum Hoffm.(Apiaceae). Acta Bot Boreal-Occident Sin. 37 (8):1540–1549.
- Liao CY, Downie SR, Li QQ, Yu Y, He XJ, Zhou B. 2013. New insights into the phylogeny of Angelica and its allies (Apiaceae) with emphasis on East Asian species, inferred from nrDNA, cpDNA, and morphological evidence. System Bot. 38(1):266–281.
- Schnittler M, Gunther KF. 1999. Central European vascular plants requiring priority conservation measures—an analysis from national Red Lists and distribution maps. Biodivers Conserv. 8(7):891–925.
- Shan RH. 1992. Umbelliferae. Vol. 55. In: Shan RH, Sheh ML, editors. Flora Reipublicae Popularis Sinicae. Beijing: Academia Sinica; p. 13–62.
- Sheh ML, Pu FT, Pan ZH, Watson MF, Cannon JFM, Holmes-Smith I, Kljuykov EV, Phillippe LR, Pimenov MG. 2005. Apiaceae. Vol. 14. In: Flora of China. St. Louis, Missouri: Missouri Botanical Garden Press; Beijing: Science Press; p. 1–205.
- Zhang SY, Liao CY, Li ML, Chen Y, Zhou B. 2018. Research on pollen morphologies of Ostericum Hoffm. (Apiaceae) of eight species from seventeen populations. Acta Bot Boreal-Occident Sin. 38 (12):2224–2235.
- Zych M, Michalska B, Krasicka-Korczyńska E. 2014. Myophyly in the critically endangered umbelliferous plant Ostericum palutre Besser (Apiaceae). Plant Syst Evol. 300(1):187–196.