Abstract
In this study, the complete mitochondrial genome of Ghauriana sinensis was sequenced and annotated. The mitogenome of G. sinensis is circular and 15,491 bp in size. The total length of 13 protein-coding genes (PCGs) is 10,942 bp, which encode 3647 amino acids. The phylogenetic tree with reference sequences confirmed that G. sinensis belonged to the Typhlocybinae but distinct from other species in this subfamily.
The leafhopper tribe Empoascini belongs to the subfamily Typhlocybinae (Hemiptera: Auchenorryncha: Cicadellidae), known as small size some 3–5 mm and rich species, especially part of them are major economic plant pest. This group species are always similar in green and yellow color shape, superadd the small size and less stripe marked, which made it difficult to be identified only basing on morphological characteristics. Therefore, it is necessary to use molecular data as a tool to explore the taxonomic status of each species in this subfamily accurately. Currently, only six complete mitochondrial genomes had been sequenced in this tribe while three of them had been published (Zhou et al. Citation2016; Liu et al. Citation2017; Luo et al. Citation2019).
The genus Ghauriana contains only two species worldwide. Ghauriana sinensis was first recorded in China by Qin et al. Citation2011. In this study, the materials of G. sinensis were collected from a hackberry tree in Jiuchang, Xiuwen, Guiyang, Guizhou, China, at June 2019 and kept in 100% anhydrous ethanol. The entire body (without abdomen) was used to be extracted the total genomic DNA by the Qiagen DNeasy kit (Venlo, the Netherlands). The male genitalia was deposited in the Institute of Entomology, Guizhou University, Guiyang, China (GUGC), accession number of them is GUGC-IDT-00190. Sequencing was conducted by Illumina NovaSeq6000 platform (Berry Genomics, Beijing, China) and the reads were assembled and annotated by using Generous Prime (v2019.1.3). All tRNA genes were identified by ARWEN v1.2 (Laslett and Canbäck Citation2008).
The circular mitogenome of G. sinensis is 15,491bp in size (GeneBank accession number: MN699874). Complete mitogenome of G. sinensis contained 13 PCGs, 22 transfer RNA genes, two ribosomal RNA genes and one A + T-rich region. The A + T content of G. sinensis mitochondrial genome is 79.6% (A: 38.7%, T:40.9%, C:10.4%, G:10%). The total length of 13 protein-coding genes (PCGs) is 10,942 bp, encoding 3,647 amino acids. All PCGs start with ATN codon, except ATP8 using TTG. Nine PCGs have TAN as the stop codon, while the COX1, COX2, COX3, and ND5 have an incomplete stop codon (T). The length of 16S rRNA and 12S rRNA are 1145 bp and 781 bp, respectively.
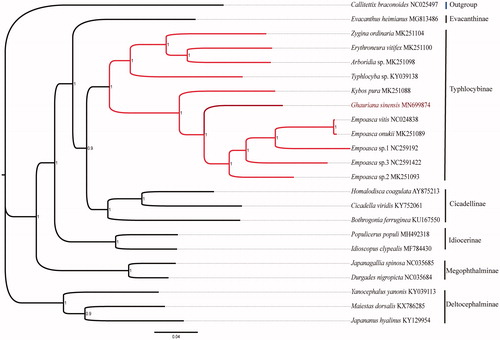
To validate the phylogenetic position, phylogenetic tree was constructed by using the nucleotide sequences of 13 PCGs, 21 reference sequences from other taxa in the family Cicadellidae, and one sequences from family Cercopidae as outgroup. For all the sequences, the third nucleotide of each codon were removed. The GTR + I + G model was determined by MrBayes3.2.7. on CIPRES platform. The phylogenetic relationship between G. sinensis and other species had been revealed. As shown in the phylogenetic tree (), species from same subfamily were clustered into one clade. It also confirmed that G. sinensis belonged to the Typhlocybinae but different from other species in the subfamily.
Acknowledgments
We sincerely appreciate Dr. Haoxi Li (College of Tobacco Science, Guizhou University, Guiyang, China) and Dr. Bin Yan (Guizhou Provincial Key Laboratory for Agricultural Pest Management of the Mountainous Region, Institute of Entomology, Guizhou University, Guiyang, China) for help with manuscript and data analysis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequence. Bioinform. 24(2):172–175.
- Liu JH, Sun CY, Long J, Guo JJ. 2017. Complete mitogenome of tea green leafhopper, Empoasca onukii (Hemiptera: Cicadellidae) from Anshun, Guizhou Province in China. Mitochondr DNA B. 2(2):808–809.
- Luo X, Chen Y, Chen C, Pu DQ, Tang XB, Zhang H, Lu DH, Mao JH. 2019. Characterization of the complete mitochondrial genome of Empoasca sp. (Cicadellidae: Hemiptera). Mitochondr Part B. 4(1):1477–1478.
- Qin DZ, Liu Y, Zhang YL. 2011. A taxonomic study of Chinese Empoascini (Hemiptera: Cicadellidae: Typhlocybinae). Zootaxa. 2923(1):48–58.
- Zhou NN, Wang MX, Cui L, Chen XX, Han BY. 2016. Complete mitochondrial genome of Empoasca vitis (Hemiptera: Cicadellidae). Mitochondr DNA A. 27(2):1052–1053.