Abstract
In the ornamental plant breeding, breeders always use distant hybridization to create new cultivars. Recent sequencing technology helps obtaining high-quality chloroplast genomes, which benefits the line breeding of plants as the super-barcodes. Herein, we established the complete chloroplast genome of Prunus campanulata ‘Fei han’ to lay a foundation in the future genetic comparison and modification. The 157,919 bp chloroplast genome presented a large single copy (LSC) region of 85,929 bp and a small single copy (SSC) region of 19,118 bp, separated by two inverted repeat (IRs) regions of 26,436 bp. The overall GC content was 36.72%, and SSC component show a lower GC percentage of 30.22, and same IR region of 42.53% G + C with other cherry cultivars. A total of 116 CDSs were found, 37 tRNA genes, and 8 rRNA genes. As expected, Prunus campanulata ‘Fei han’ was close to Prunus campanulata within the subsection Cerasus from phylogenetic analysis This chloroplast genome announcement of the Prunus campanulata ‘Fei han’ could embody light on the artificial breeding in the Cerasus with luxuriant genetic messages.
Prunus campanulata is one of the most charming cherry original species under the Rosaceae order. Therefore, many breeders use P. campanulata as the crossing parent in the modern breeding to create new ornamental cultivars. Prunus campanulata ‘Fei han’ is one of the excellent offsprings from the hybridizations of P. campanulata. This cultivar exhibits intermediate characters from its parents with light pink petal color. Due to its earlier flowering date in spring, it is always planted with Cerasus × yedoensis, in which the landscape can form a continuous flowering scenery. The tolerance to cold and drought of ‘Fei han’ is stronger than that of common cherry trees, especially under severe weather. P. campanulata ‘Fei han’ is a potential parent in further selection and hybridization breeding. However, related genetic information and relationship based on it have not been well established (Zhang et al. Citation2018). Now the rapid sequencing technology promotes the acquisition of abundant genetic information for phylogenetic research and genetic engineering (Shirasawa et al. Citation2019), including assembling a high-precision chloroplast. Thus, we reported the high-quality chloroplast genome of Prunus campanulata ‘Fei han’ to lay the foundation in the genetic recognition and functional analysis. The comparisons of related species in same genus also provided a verification for the application of super barcode in the cherry cultivars natural and artificial evolution in genus Cerasus (Mu et al. Citation2018).
Leaves of P. campanulata ‘Fei han’ were collected from the cherry breeding farm in Fuzhou (location: 26°04’51.3”N 119°14’19.9”E) and samples were preserved in Fujian Agriculture and Forestry University. Total genomic DNA was extracted by modified CTAB method to avoid the influence of high polysaccharides and phenols. The frozen samples including fresh tissues, specimens and sequenced DNA can be found in the laboratory of Fujian Agriculture and Forestry University (Voucher specimen: YT-FJ2019-4A, FAFU, 23°32′25.19″N 120°47′57.71″E). PE150 library strategy were adopted and raw sequences were obtained from the BGI-500 high-throughput platform (BGI, Wuhan, China) (Mak et al. Citation2017). We obtained total about 6 Gb clean reads after removing adapters and low-quality reads by fastp software (Chen et al. Citation2018). Then the processed data were assembled by GetOrganelle v1.5.2 flow, in which core mapping software and assembly tool were bowtie2 and Spades version 3.13.1 (Vasilinetc et al. Citation2015). Random separated reads were then rectified and assembled into contigs. Fragments with low sequence coverages were removed as noises during the screening progress by using Bandage v0.8.1 (Wick et al. Citation2015) and ultimately formed a high-coverage circle chloroplast. Then, clean reads were mapped to the draft genome to check the assembling consistency. Detailed basic information was counted by Bioedit. The genome was preliminarily annotated for coding genes and RNA using Geneious Prime to adjust the starting position. As a result, we established a length of 157,919 bp circle chloroplast genome of P. campanulata ‘Fei han’ with a total GC content of 36.72%. This cp genome typical includes a length of 85,929 bp large single-copy (LSC) region and a 19,118 bp small single-copy (SSC) region, separated by two 26,436 bp inverted repeat (IRs). The four parts manifested an unbalanced GC contents and among the nearby cultivars there are fewer differences in each structure. LSC and SSC, 34.59% and 30.22%, are interrupted by two 42.53% GC content IRs from both sides. After assessment of the assembled plastid genome, annotation of the new plastid genome was conducted by online software GeSeq. In this chloroplast genome, 37 tRNA and 8 conserved rRNA were found respectively. 116 CDSs were annotated by hmmscan. The assembled cp genome sequence of P. campanulata ‘Fei han’ can be detected in GenBank with an accession number of MT018451.
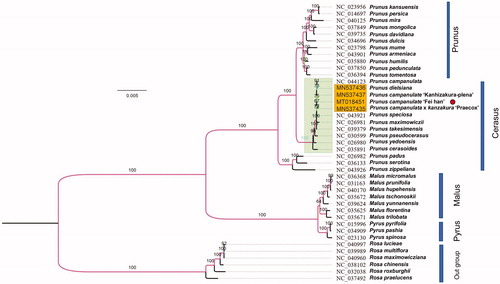
To obtain a clear phylogenetic position of the newly sequenced cultivar, P. campanulata ‘Fei han’ was clustered along with most representative Prunus species and part related chloroplast genomes. All the cp genomes were aligned following HomBlocks pipeline (Bi et al. Citation2018). We reconstructed a phylogeny employing the GTRCAT model and 1000 bootstrap replicates under the maximum-likelihood (ML) inference by means of RAxML-HPC v.8.2.10 on the CIPRES cluster. The new ML tree () was consistent with recent phylogenetic results on Prunus, species that Rosa genus located was chosen as outgroup taxa. P. campanulata ‘Fei han’ displayed a closer kinship to Prunus campanulata and Prunus speciose within the subsection Cerasus. The whole cluster used in Rosaceae fell into four main branches, which may indicate Prunus endured a separation from Cerasus genus. The cultivars from P. campanulata gathered in same branch with certain differences. The super-barcode may contribute to the endogenous recognitions in the further applications of plant cultivars. We believe the presentation of P. campanulata ‘Fei han’ chloroplast genome helps clarify its evolutionary status in genus Prunus especially among nearby cultivars, and provides genomic resources for artificial breeding and fundamental researches.
Figure 1. Maximum-likelihood (ML) phylogenetic tree of selected chloroplast sequences in Rosaceae. The branch that contain six species in Rosa genus is treated as the outgroup. Prunus campanulate ‘Fei han’ is marked with red circle. Genebank accession numbers were listed before their corresponding species.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: a multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Mak SST, Gopalakrishnan S, Caroe C, Geng C, Liu S, Sinding MS, Kuderna LFK, Zhang W, Fu S, Vieira FG, et al. 2017. Comparative performance of the BGISEQ-500 vs Illumina HiSeq2500 sequencing platforms for palaeogenomic sequencing. Gigascience. 6(8):1–13.
- Mu X, Wang P, Du J, Gao YG, Zhang J. 2018. The chloroplast genome of Cerasus humilis: Genomic characterization and phylogenetic analysis. PLoS One. 13(4):e0196473.
- Shirasawa K, Esumi T, Hirakawa H, Tanaka H, Itai A, Ghelfi A, Nagasaki H, Isobe S. 2019. Phased genome sequence of an interspecific hybrid flowering cherry, ‘Somei-Yoshino’ (Cerasus × yedoensis). DNA Res. 26(5):379–389. doi:10.1093/dnares/dsz016
- Vasilinetc I, Prjibelski AD, Gurevich A, Korobeynikov A, Pevzner PA. 2015. Assembling short reads from jumping libraries with large insert sizes. Bioinformatics. 31(20):3262–3268.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352.
- Zhang J, Chen T, Wang Y, Chen Q, Sun B, Luo Y, Zhang Y, Tang H, Wang X. 2018. Genetic Diversity and Domestication Footprints of Chinese Cherry [Cerasus pseudocerasus (Lindl.) G.Don] as Revealed by Nuclear Microsatellites. Front Plant Sci. 9:238.
