Abstract
Oxybasis glauca (L.) S. Fuentes, Uotila & Borsch is an annual halophyte. In this study, we presented complete chloroplast genome of O. glauca as the first chloroplast genome of genus Oxybasis, which is 151,655 bp long and has four subregions: 83,759 bp of large single-copy (LSC) and 17,914 bp of small single-copy (SSC) regions are separated by 24,991 bp of inverted repeat (IRs) regions including 129 genes (84 protein-coding genes, 8 rRNAs, and 37 tRNAs). The overall GC content of the chloroplast genome is 36.9% and those in the LSC, SSC, and IR regions are 34.7, 30.4, and 42.8%, respectively. Phylogenomic tree shows that O. glauca belongs to tribe Atripliceae forming a monophyletic clade with genus Chenopodium and Atriplex.
Oxybasis glauca (L.) S. Fuentes, Uotila & Borsch is an annual halophyte that is widely distributed in most continents as an alien species. Because of salinity tolerance, this species has been investigated for identifying factors of signaling pathway related to seeds germination under salinity conditions (Duan et al. Citation2004; Chen et al. Citation2012; Wang et al. Citation2017). In recent phylogenetic studies, O. glauca were treated as genus Oxybasis from genus Chenopodium and supported by several follow-up studies (Fuentes-Bazan, Uotila, et al. Citation2012; Kolano et al. Citation2015; Kadereit et al. Citation2017). Since O. glauca displays continuous morphological variations with a wide geographic distribution, Mosyakin (Citation2013) considers it as species aggregate rather than single species. Additional molecular study of this species complex is strongly required even though its taxonomic treatments (Mosyakin Citation2013; Verloove Citation2013; Mosyakin and De Lange Citation2018), indicating that complete chloroplast genomes can provide both high resolution of phylogenetic studies and basis for developing efficient molecular markers with previously deciphered chloroplast genome of neighbor genera.
We completed chloroplast genome of O. glauca isolated in Buan-gun, Jeollabuk-do, Korea (Voucher in InfoBoss Cyber Herbarium (IN); IB-01023). Total DNA of O. glauca was extracted from fresh leaves by using a DNeasy Plant Mini Kit (QIAGEN, Hilden, Germany). Genome sequencing was performed using HiSeqX at Macrogen Inc., Korea. de novo assembly was done by Velvet 1.2.10 (Zerbino and Birney Citation2008) and SOAPGapCloser 1.12 (Zhao et al. Citation2011) and confirmed based on alignments by BWA 0.7.17 (Li Citation2013) and SAMtools 1.9 (Li et al. Citation2009). Geneious R11 11.1.5 (Biomatters Ltd., Auckland, New Zealand) was used for chloroplast genome annotation based on C. ficifolium chloroplast complete genome (NC_041200; Kim et al. Citation2019).
The chloroplast genome of O. glauca (GenBank accession is MK814481) is 151,655 bp (GC ratio is 36.9%) and has four subregions: 83,759 bp of large single-copy (34.7%) and 17,914 bp of small single-copy (30.4%) regions are separated by 24,991 bp of inverted repeat (IRs; 42.8%). It contains 129 genes (84 protein-coding genes, 8 rRNAs, and 37 tRNAs); 16 genes (five protein-coding genes, four rRNAs, and seven tRNAs) are duplicated in IR regions.
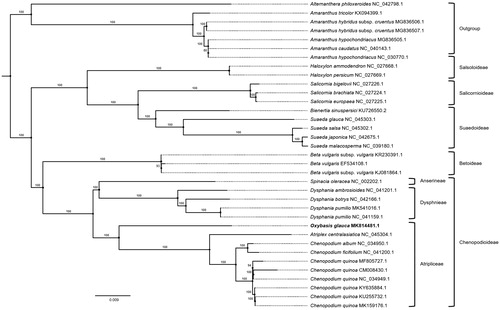
Thirty-five complete chloroplast genomes of Amaranthaceae were used for constructing phylogenic trees. Whole chloroplast genome sequences were aligned by MAFFT 7.450 (Katoh and Standley Citation2013) for bootstrapped constructing maximum likelihood trees using IQ-TREE 1.6.12 (Nguyen et al. Citation2015). Oxybasis glauca belongs to tribe Atripliceae with genus Chenopodium and Atriplex like the previous studies (Fuentes-Bazan, Mansion, et al. Citation2012; Fuentes-Bazan, Uotila, et al. Citation2012; Kolano et al. Citation2015; Kadereit et al. Citation2017). The four tribes (sensu Fuentes-Bazan, Uotila, et al. Citation2012) in Chenopodioideae are reciprocally monophyletic; but their phylogenic relationship is unclear due to low resolution (Fuentes-Bazan, Uotila, et al. Citation2012) or topological incongruence between nuclear and chloroplast trees (Fuentes-Bazan, Mansion, et al. Citation2012; Fuentes-Bazan, Uotila, et al. Citation2012). Our tree shows phylogenic relationship in Chenopodioideae with high resolution but it requires additional chloroplast genomes of tribe Axyrideae which was not included in this study to fully understand the phylogenic relationship of Chenopodioideae .
Figure 1. Maximum likelihood (bootstrap repeat is 1000) phylogenetic tree of 35 Amaranthaceae complete chloroplast genomes: Oxybasis glauca (MK814481.1, in this study), Alternanthera philoxeroides (NC_042798.1), Amaranthus tricolor (KX094399.1), Amaranthus hybridus subsp. cruentus (MG836506.1), Amaranthus hypochondriacus (MG836505.1), Amaranthus caudatus (NC_040143.1), Amaranthus hypochondriacus (NC_030770.1), Haloxylon ammodendron (NC_027668.1), Haloxylon persicum (NC_027669.1), Salicornia bigelovii (NC_027226.1), Salicornia brachiata (NC_027224.1), Salicornia europaea (NC_027225.1), Bienertia sinuspersici (KU726550.2), Suaeda glauca (NC_045303.1), Suaeda salsa (NC_045302.1), Suaeda japonica (NC_042675.1), Suaeda malacosperma (NC_039180.1), Beta vulgaris subsp. vulgaris (KR230391.1), Beta vulgaris (EF534108.1), Beta vulgaris subsp. vulgaris (KJ081864.1), Spinacia oleracea (NC_002202.1), Dysphania ambrosioides (NC_041201.1), Dysphania botrys (NC_042166.1), Dysphania pumilio (MK541016.1), Dysphania pumilio (NC_041159.1), Atriplex centralasiatica (NC_045304.1), Chenopodium album (NC_034950.1), Chenopodium ficifolium (NC_041200.1), Chenopodium quinoa (MF805727.1), Chenopodium quinoa (CM008430.1), Chenopodium quinoa (NC_034949.1), Chenopodium quinoa (KY635884.1), Chenopodium quinoa (KU255732.1), Chenopodium quinoa (MK159176.1). The numbers above branches indicate bootstrap support values of maximum likelihood phylogenetic tree.
Disclosure statement
No conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen S, Xing J, Lan H. 2012. Comparative effects of neutral salt and alkaline salt stress on seed germination, early seedling growth and physiological response of a halophyte species Chenopodium glaucum. Afr J Biotechnol. 11(40):9572–9581.
- Duan D, Liu X, Khan MA, Gul B. 2004. Effects of salt and water stress on the germination of Chenopodium glaucum L. seed. Pak J Bot. 36(4):793–800.
- Fuentes-Bazan S, Mansion G, Borsch T. 2012. Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae). Mol Phylogenet Evol. 62(1):359–374.
- Fuentes-Bazan S, Uotila P, Borsch T. 2012. A novel phylogeny-based generic classification for Chenopodium sensu lato, and a tribal rearrangement of Chenopodioideae (Chenopodiaceae). Willdenowia. 42(1):5–24.
- Kadereit G, Newton RJ, Vandelook F. 2017. Evolutionary ecology of fast seed germination—a case study in Amaranthaceae/Chenopodiaceae. Perspect Plant Ecol Evol Systemat. 29:1–11.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kim Y, Chung Y, Park J. 2019. The complete chloroplast genome of Chenopodium ficifolium Sm.(Amaranthaceae). Mitochondr DNA B. 4(1):872–873.
- Kolano B, Siwinska D, McCann J, Weiss-Schneeweiss H. 2015. The evolution of genome size and rDNA in diploid species of Chenopodium sl (Amaranthaceae). Bot J Linn Soc. 179(2):218–235.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv.:1303.3997.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079.
- Mosyakin SL. 2013. New nomenclatural combinations in Blitum, Oxybasis, Chenopodiastrum, and Lipandra (Chenopodiaceae). Phytoneuron. 56:1–8.
- Mosyakin SL, De Lange PJ. 2018. New combinations for three taxa of the Oxybasis glauca aggregate (Chenopodiaceae) from Australasia, East Asia, and South America. Phytotaxa. 350(3):259–273.
- Nguyen L-T, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Verloove F. 2013. A new combination in Oxybasis (Amaranthaceae). New J Bot. 3(1):59–60.
- Wang J, Cheng G, Wang C, He Z, Lan X, Zhang S, Lan H. 2017. The bHLH transcription factor CgbHLH001 is a potential interaction partner of CDPK in halophyte Chenopodium glaucum. Sci Rep. 7(1):1–16.
- Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18(5):821–829.
- Zhao Q-Y, Wang Y, Kong Y-M, Luo D, Li X, Hao P. 2011. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinf. 12(Suppl 14):S2.
