Abstract
Enhalus acoroides is the largest tropic seagrass, occurring in the shallow coastal areas. In this study, we characterized the complete chloroplast genome sequence of this species. The whole chloroplast genome was 176,211 bp in length with a typical quadripartite structure, including one large single-copy (LSC) region (89,851 bp), one small single-copy (SSC) region (2150 bp), and a pair of inverted repeats (IRs) (42,105 bp). The guanine-cytosine (GC) content of this genome was 38.4%. A total of 142 genes were found in this genome, including 95 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. The phylogenetic analysis indicated that E. acoroides and Thalassia hemprichii formed a distinct clade.
Enhalus acoroides, the sole species of Enhalus, is widely distributed in the tropical shallow intertidal areas of the Indian Ocean and the western Pacific Ocean (Short et al. Citation2007). In China, E. acoroides is the dominant species along the east coastline of Hainan Island, of which the distribution area has been declining dramatically (Chen et al. Citation2015). This dioecious seagrass displays the capacity for both sexual and clonal reproduction (den Hartog Citation1970; Yu et al. Citation2019). Most of the seagrasses pollinate underwater mediated by water currents (Ackerman Citation2006). However, E. acoroides is the only seagrass species that pollinate on the water surface. The small mature male flowers float to the water surface and then release pollen grains for pollination, which is similar to the freshwater aquatics Vallisneria (Cox Citation1988). In this study, we sequenced the complete chloroplast genome of E. acoroides using next-generation technology. This genome will be very useful for the phylogenetic studies of seagrasses.
Fresh plant of E. acoroides was collected from Xincun Bay, Hainan Province, China (18.41°N, 109.99°E) and the specimen was stored at Fourth Institute of Oceanography Herbarium (XC201908-2). We collected a piece of young leaf and washed it with fresh water. The genomic DNA was extracted according to a modified CTAB method and sequenced using the Illumina Novaseq platform. Low-quality reads and adapters were removed by the FastQC software (Andrews Citation2010). De novo genome assembly was conducted by SPAdes v3.9 (Bankevich et al. Citation2012). We annotated the complete chloroplast genome by DOGMA (Wyman et al. Citation2004). The annotations of tRNA genes were performed by ARAGORN (Laslett and Canback Citation2004). The complete chloroplast genome of E. acoroides was submitted to GenBank database (Accession Number: MN736636).
The complete chloroplast genome of E. acoroides was 176,211 bp in length with a typical circle structure, including one large single-copy region (LSC: 89,851 bp), one small single-copy region (SSC: 2150 bp), and a pair of inverted repeats (IRs: 42,105 bp). The guanine-cytosine (GC)-content was 38.4%. There were a total of 142 genes in this genome, consisting of 95 protein-coding genes, 39 tRNA genes, and 8 rRNA genes. Most of the protein-coding genes located in the LSC and IRS regions. The short small single-copy region was very short, only containing two genes (ndhD and psaC).
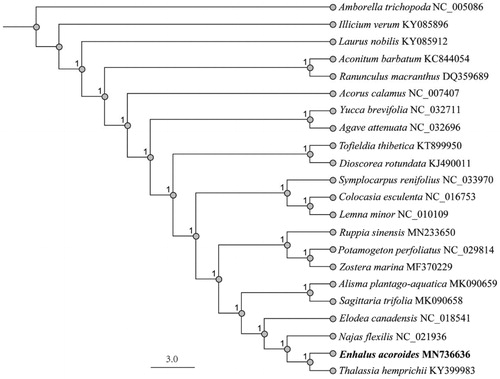
To clarify the phylogenetic position of E. acoroides, we downloaded 21 completed chloroplast genomes from GenBank database. The phylogenetic trees were analyzed with MEGA6 software (Tamura et al. Citation2013) using maximum likelihood (ML) method. Bootstrap values were calculated out of 1000 replicates. As shown in the phylogenetic tree (), E. acoroides formed a distinct clade with Thalassia hemprichii.
Geolocation information
Xincun Bay, Hainan Province, China (18.41°N, 109.99°E).
Acknowledgements
We thank Ruiting Gu for comments on this paper.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Ackerman JD. 2006. Sexual reproduction of seagrasses: Pollination in the marine context. In: Larkum, AWD, Orth RJ, Duarte CM, editors. Seagrasses, biology, ecology and conservation. Dordrecht, The Netherlands: Springer; p. 89–109.
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Cambridge (UK): Babraham Institute.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chen SQ, Wang DR, Wu ZJ, et al. 2015. Discussion of the change trend of the seagrass beds in the east coast of Hainan Island in nearly a decade. Marine Environ Sci. 37:106–113. (In Chinese).
- Cox PA. 1988. Hydrophilous pollination. Annu Rev Ecol Syst. 19(1):261–279.
- Den Hartog C. 1970. The sea-grasses of the world. Amsterdam, The Netherlands: North Holland Publishing Company.
- Laslett D, Canback B. 2004. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 32(1):11–16.
- Short F, Carruthers T, Dennison W, Waycott M. 2007. Global seagrass distribution and diversity: a bioregional model. J Exp Mar Biol Ecol. 350(1-2):3–20.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Yu S, Wu Y, Serrao EA, Zhang J, Jiang Z, Huang C, Cui L, Thorhaug A, Huang X. 2019. Fine-scale genetic structure and flowering output of the seagrass Enhalus acoroides undergoing disturbance. Ecol Evol. 9(9):5186–5195.