Abstract
The complete plastid genome of Stenotaphrum subulatum Trin. was determined and analyzed in this work. The plastid genome was 139,282 bp in length with 82,220 bp of the large single-copy (LSC) region, 12,392 bp of the small single-copy (SSC) region, and 22,335 bp of the invert repeats (IR) regions. The genome contained 136 genes, 87 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. The maximum likelihood(ML)phylogenetic tree suggested that S. subulatum Trin. is closely to S. secundatum.
Stenotaphrum subulatum Trin. (Panicoideae) is a widely used warm season turf grass for home lawns and commercial landscapes (Sauer Citation1972). It grows on 0–1100 m altitudes and are widely distributed in the tropical and subtropical areas of China (Chen and Phillips Citation2006; Liu Citation2010). It has characteristics of broad leaf blades and rapid stolon production and is widely used in home lawns, recreation parks, and sports fields (Wang et al. Citation2017). The plastid genome is valuable in plant systematics research due to its highly conserved structures, uniparental inheritance, and haploid nature (Fu et al. Citation2016). Plastid genome have also been smartly engineered to confer useful agronomic traits and/or serve as bioreactors (Jin and Daniell Citation2015). As a high quality turf grass, the complete plastid genome of S.subulatum Trin. was sequenced, which will provide genomic and genetic sources for further research.
In this study, fresh leaf sample of S.subulatum Trin. was acquired from Money Island (16°26′N, 111°30′E), Sansha, Hainan Province of China. The voucher specimen was kept in Key Laboratory of Genetics and Germplasm Innovation of Tropical Special Forest Trees and Ornamental Plants, Hainan University (HD20191212). Total DNA of S. subulatum Trin. was extracted from the fresh, young leaves (about 1.5 g) with a modified CTAB method (Doyle and Doyle Citation1987), with 400 bp randomly interrupted for library construction. Approximately, 5 Gb of sequences data were extracted from the total sequencing output and input into Organelle PBA (Soorni et al. Citation2017) to assemble the plastid genome. Annotation of the plastid genome was performed using the Dual Organellar GenoMe Annotator (DOGMA) online tool (Wyman et al. Citation2004) and Geneious v11.1.5 (Kearse et al. Citation2012), then manually verified and corrected by comparison with S.secundatum (NC_036704.1). Finally, we obtained a complete plastid genome of S.subulatum Trin. and submitted to GenBank with accession number (MN989414).
The complete plastid genome sequence of S.subulatum Trin. is 139,282 bp in length with 82,220 bp of the large single-copy (LSC) region, 12,392 bp of the small single-copy (SSC) region and 22,335 bp of the invert repeats (IR) regions. The overall GC content of the plastid genome was 38.54%, while the corresponding values of the LSC, SSC, and IR regions were 36.37, 33.01, and 44.05%, respectively. The genome contained 136 genes, 87 protein-coding genes, 41 tRNA genes, and 8 rRNA genes. Among those, 8 protein-coding genes (ndhB, rpl2, rpl23, rps7, rps15, rps19, ycf15, and ycf2), 8 tRNA genes(trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC), and 4 rRNA genes were duplicated in IR regions. Fourteen genes (8 protein-coding genes and 6 tRNA genes) contained one intron, and two genes (rps12 and ycf3) contained two introns.
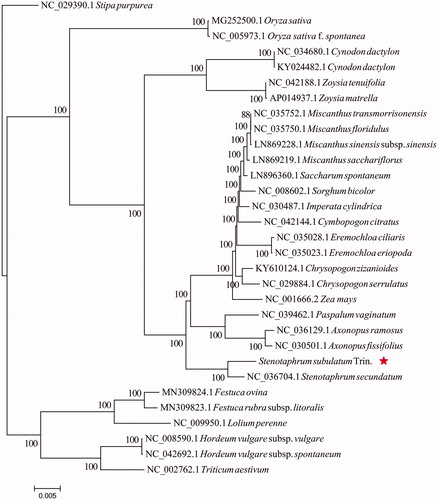
The maximum likelihood phylogenetic tree was generated based on the plastid genome of S. secundatum and other 30 species of the Poaceae. Alignment was conducted using MAFFT v7.307 (Katoh and Standley, Citation2013). The phylogenetic tree was built using RAxML (Stamatakis Citation2014) with bootstrap set to 1000. Stipa purpurea (NC_029390.1) served as the outgroup. The maximum likelihood(ML)phylogenetic tree suggested that S. subulatum Trin. is closely related to S. secundatum (). The complete plastid genome sequence of S. subulatum Trin. will provide genetic and genomic information to promote its allies in the future.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen SL, Phillips SM. 2006. The flora of China. , Beijing, China: Science Press.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Fu PC, Zhang YZ, Geng HM, Chen SL. 2016. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ. 4:e2540.
- Jin S, Daniell H. 2015. The engineered chloroplast genome just got smarter. Trends Plant Sci. 20(10):622–640.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Liu GD. 2010. Panicoideae of Hainan. Beijing, China: Science Press.
- Sauer JD. 1972. Revision of Stenotaphrum (Panicoideae: Paniceae) with attention to its historical geography. Brittonia. 24(2):202–222.
- Soorni A, Haak D, Zaitlin D, Bombarely A. 2017. Organelle_PBA, a pipeline for assembling chloroplast and mitochondrial genomes from PacBio DNA sequencing data. BMC Genomics. 18(1):49.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang ZY, Raymer P, Chen ZB. 2017. Isolation and Characterization of Microsatellite Markers for Stenotaphrum Trin. Using 454 Sequencing Technology. Horts. 52 (1):16–19.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.