Abstract
Anoectochilus nandanensis and A. calcareus are medical plants with important economic value. In this study, we assembled their complete chloroplast (cp) genome sequences. The assembled cp genomes are 152,415 bp and 151,864 bp in length, including a large single-copy (LSC) region of 82,412 bp and 82,083 bp, a small single-copy (SSC) region of 17,367 bp and 17,142 bp and a pair of inverted repeats (IRs) of 26,318 bp and 26,320 bp, respectively. All genomes contained 133 genes, including 79 protein-coding genes, 30 tRNA genes, and 4 rRNA genes. The overall GC content of genomes in both species is 36.9%. Further phylogenetic analysis showed that three Anoectochilus comprised of a monophyletic clade and A. nandanensis was more close to A. emeiensis.
The Anoectochilus genus in Orchidaceae includes about 40 species (Editorial Committee of Flora of China Citation2009; Qu et al. Citation2015), in which Anoectochilus calcareus Aver. and Anoectochilus nandanensis Y. Feng Huang & X. C. Qu are local medical plants with important economic value. However, chloroplast sequence variations of these two species remain unclear. In this study, complete chloroplast genomes of A. nandanensis and A. calcareus were sequenced and analyzed for improving the understanding of their genetic variations, which will serve effective conservation and utilization of its genetic resources. Each sample of A. nandanensis and A. calcareus were collected from fresh leaves of only one individual. Voucher specimens (Yunfeng Huang 201901113, Yunfeng Huang 201901114) were collected from Guangxi for A. nandanensis (N24°59′7.44″, E107°56′28.68″, 640 m) and A. calcareus (N23°0′19.44″, E105°51′55.54″, 1113 m) and deposited in the Herbarium of Guangxi Institute of Traditional Medical and Pharmaceutical Science (GXMI). The total DNA was extracted using the modified CTAB method (Doyle Citation1987). DNA data were sequenced using the Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA). Subsequently, chloroplast genome assembly using GetOrganelle (Jin et al. Citation2018) and the annotation was performed with PGA (Qu et al. Citation2019), then confirmed manually. Finally, the complete chloroplast genome was deposited in GenBank (accessible numbers: MT021458 and MT021459).
The complete chloroplast genomes of A. nandanensis and A. calcareus were 152,415 bp and 151,864 bp in length, respectively. They exhibit the typical quadripartite structure, including a pair of IRs (26,318 bp and 26,320 bp, respectively) separated by the LSC (82,412 bp and 82,083 bp, respectively) and SSC (17,367 bp and 17,142 bp, respectively) regions. Each of them contains 113 unique genes, including 79 protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. Twenty genes were duplicated in the IRs, giving a total of 133 genes. Overall, GC contents of the whole chloroplast genomes from both A. nandanensis and A. calcareus are 36.9%.
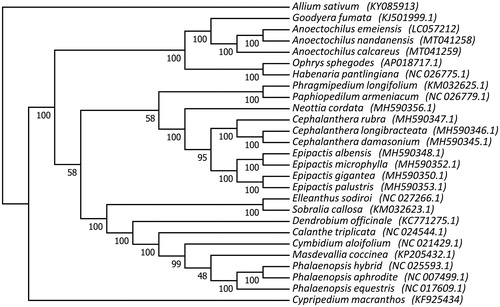
In order to clarify their phylogenetic relationships, we further used the software RAxML to construct a phylogenetic tree with 1000 bootstraps under the GTRGAMMA substitution mode (Stamatakis Citation2014). Twenty six complete chloroplast genomes in Orchidaceae and one outgroup were selected in this study, including three Anoectochilus species. All these sequences were aligned using the software MAFFT v.7 (Katoh and Standley Citation2013). Our phylogenetic analysis showed that three Anoectochilus species comprised of a monophyletic clade and A. nandanensis was more close to A. emeiensis .
Disclosure statement
No potential conflict of interest is reported by the author(s).
Additional information
Funding
References
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Editorial Committee of Flora of China. 2009. Flora of China. Vol. 25. Beijing (China): Science Press.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. BioRxiv.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Qu XC, Huang YF, Feng HZ, Hu RC. 2015. Anoectochilus nandanensis, sp. nov. (Orchidaceae) from Northern Guangxi, China. Nord.J Bot. 33(5):572–575.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15:50.
- Stamatakis A. 2014. RAxML version 8.2.12: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.