Abstract
The domestic South American camelid Vicugna pacos L. is distributed along Peru, Chile, Bolivia, and Argentina. Here, we contribute to the bioinformatics and evolutionary systematics of the Camelidae by performing high-throughput sequencing analysis on the black Huacaya breed of V. pacos from Puno, Peru. The black Huacaya breed mitogenome is 16,664 base pairs (bp) in length and contains 37 genes (GenBank accession MT044302). The mitogenome shares a high-level of gene synteny to other Camelidae (Camelops, Camelus, Lama, and Vicugna). The mitogenome of the black Huacaya breed of V. pacos situates it in a clade with V. vicugna Molina, sister to Lama. We anticipate that further mitogenome sequencing of different breeds from Vicugna pacos will improve our understanding of the evolutionary history of this taxon.
Alpacas (Vicugna pacos) and llamas (Lama glama L.), domestic South American camelids, are the basis for livestock production in the High-Andean zones of Peru (Paredes et al. Citation2014). More than four million of alpacas in Peru positioned it as the first camelids fiber producer worldwide (90% of world production, Paredes et al. Citation2013). This alpaca population is composed of the Huacaya (more than 85%) and Suri breed (Quispe et al. Citation2009, Paredes-Peralta et al. Citation2011). Although selection pressures during many generations for fiber and color traits possibly lead to the loss of genetic variability mainly in the Huacaya alpaca breeds (Presciuttini et al. Citation2010, Paredes et al. Citation2013), there is limited genomic information about their genetic differences. To contribute to the evolutionary systematics of the Camelidae and to advance the understanding of the taxonomy of the black Huacaya alpaca breed, this study characterized the complete mitochondrial genome of a male specimen of Vicugna pacos from Quimsachata Germplasm Conservation Center at the Illpa, Puno, Peru (15°47′43″S, 70°37′22″W).
DNA was extracted from blood of the black Huacaya alpaca breed (Specimen Voucher: INIA150129) using the Quick-DNA Plant/Seed kit (Zymo Research, California, USA) following the manufacturer’s instructions. The 150 bp PE Illumina library construction and sequencing was performed by myGenomics, LLC (Alpharetta, Georgia, USA). The genomes were assembled using default de novo settings in MEGAHIT (Li et al. Citation2016) and Sanger sequencing to close the gap in the control region using primers 16,107 F 5′- CCCGCATCATACAACCATAAGG-3′ and 48 R 5′-CCATCTAGGCATTTTCAGCGC-3′ following the protocol of Bustamante et al. (Citation2017). The mitogenome was confirmed using default mapping settings in Geneious Prime (Biomatters, Ltd, Auckland, New Zealand). The genes were annotated with MITOS (Bernt et al. Citation2013) and manually using ORFfinder. The Huacaya breed mitogenome was aligned to other mitogenomes using MAFFT (Katoh and Standley Citation2013). The phylogenetic analysis was executed with RAxML-NG (Kozlov et al. Citation2018) with the GTR + gamma model and 1000 bootstraps. The tree was visualized with TreeDyn 198.3 at Phylogeny.fr (Dereeper et al. Citation2008).
The mitogenome of the black Huacaya alpaca breed is 16,664 bp in length and contains 37 genes. It has a slight A + T skewed (59.1%) and includes 22 tRNA (tRNA-Leu and tRNA-Ser occur in duplicate), 2 rRNA (rnl, rns), 13 genes involved in electron transport and oxidative phosphorylation, and 1 control region (CR). The mitogenome of the Huacaya breed is similar in length, content, and organization to other 14 Camelidae belonging to the genera Camelops Leidy, Camelus L., Lama, and Vicugna (Di Rocco et al. Citation2010, Westbury et al. Citation2016).
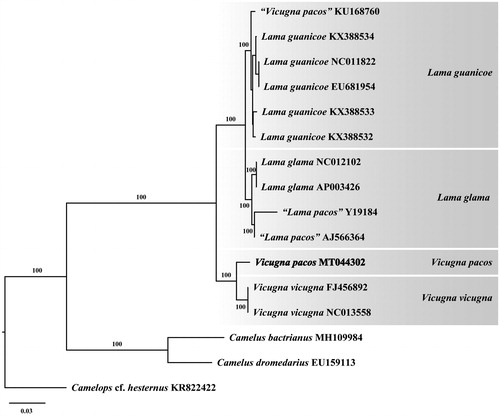
Phylogenetic analysis of the black Huacaya breed of V. pacos resolved it in a fully supported clade with V. vicugna, sister in position to the genus Lama (). A similar evolutionary relationship for Camelidae was reported by Westbury et al. (Citation2016) and Díaz-Maroto et al. (Citation2019) based on mitogenome data. Further complete mitogenome sequencing of different breeds from Vicugna pacos (i.e., Suri breed) will help improve our understanding of the phylogenetics of the South American camelids.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bustamante DE, Won BY, Miller KA, Cho TO. 2017. Wilsonosiphonia gen. nov. (Rhodomelaceae, Rhodophyta) based on molecular and morpho-anatomical characters. J Phycol. 53(2):368–380.
- Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server):W465–W469.
- Di Rocco F, Zambelli A, Mate L, Vidal-Rioja L. 2010. The complete mitochondrial DNA sequence of the guanaco (Lama guanicoe): comparative analysis with the vicuña (Vicugna vicugna) genome. Genetica. 138(8):813–818.
- Díaz-Maroto PF, Hansen AJ, Rey-Iglesia A. 2019. The first complete mitochondrial genome of an ancient South American vicuña, Vicugna vicugna, from Tulán-54 (3200–2400 B.P. Northern Chile). Mitochondrial DNA B. 4(1):340–341.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2018. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. BioRxiv. 447110.
- Li D, Luo R, Liu CM, Leung CM, Ting HF, Sadakane K, Yamashita H, Lam TW. 2016. MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 102:3–11.
- Paredes MM, Membrillo A, Azor PJ, Machaca JE, Torres D, Muñoz-Serrano A. 2013. Genetic and phenotypic variation in five populations of Huacaya Alpacas (Vicugna pacos) from Peru. Small Rumin Res. 111(1–3):31–40.
- Paredes MM, Membrillo A, Gutiérrez JP, Cervantes I, Azor PJ, Morante R, Alonso-Moraga A, Molina A, Muñoz-Serrano A. 2014. Association of microsatellite markers with fiber diameter trait in Peruvian alpacas (Vicugna pacos). Livest Sci. 161:6–16.
- Paredes-Peralta MM, Alonso-Moraga A, Analla M, Machaca-Centty J, Muñoz-Serrano A. 2011. Genetic parameters and fixed effects estimation for fibre traits in alpaca Huacaya (Lama pacos). J Anim Veterin Adv. 10:1484–1487.
- Presciuttini S, Valbonesi A, Apaza N, Antonini M, Huanca T, Renieri C. 2010. Fleece variation in alpaca (Vicugna pacos): a two-locus model for the Suri/Huacaya phenotype. BMC Genet. 11(1):70.
- Quispe EC, Rodríguez TC, Iñiguez LR, Mueller JP. 2009. Producción de fibra de alpaca, llama, vicuña y guanaco en Sudamérica. Anim Genet Resour Inf. 45:1–14.
- Westbury M, Prost S, Seelenfreund A, Ramírez JM, Matisoo-Smith EA, Knapp M. 2016. First complete mitochondrial genome data from ancient South American camelids - the mystery of the chilihueques from Isla Mocha (Chile). Sci Rep. 6(1):38708.