Abstract
Acrostichum genus is the only mangrove fern that distribute in the intertidal zones of tropical and subtropical regions. A. speciosum is one rare species occurring in only few locations in Hainan and Guangdong, China. To study the evolutionary history of fern will improve our understanding of the origin of vascular plants. Here, we provide the complete chloroplast genome of A. speciosum, which is 156,095 bp in length with a GC content of 38.45%. A pair of identical inverted repeat regions (IRs) of 24,938 bp separate a large single copy (LSC) region of 84,476 and a small single copy (SSC) region of 21,744 bp. A total of 112 genes were annotated, including 72 protein-coding genes, 32 tRNA genes, and 8 rRNA genes. Phylogenetic analysis based on 30 ferns showed that A. speciosum and Ceratopteris richardii, both are aquatic plants, formed an original base clade in Pteridaceae. Our work reports the complete chloroplast genome of mangrove fern A. speciosum, which will help to improve understanding of the evolutionary history of ferns.
Mangroves contain 84 species from about 16 families, which inhabit in the intertidal ecosystems of tropical to subtropical coasts (Nabeelah et al. Citation2019). There exist three fern species of mangroves, Acrostichum speciosum, Acrostichum aureum and a hybrid Acrostichum aureum x Acrostichum speciosum (Yamamoto et al. Citation2016). To investigate the evolutionary history of fern plants will greatly improve our understanding of the origin of vascular plants. Plastid genomes display remarkable conservation in sequence and stability in gene organization, making them ideal molecules to study plant phylogeny (Daniell et al. Citation2016). Unlike the well-studied seed plants, chloroplast genome investigations of ferns are limited (Robison et al. Citation2018). To the aspect of mangrove ferns, chloroplast genes were used as the molecular marker for hybrid identification (Zhang et al. Citation2013). In addition, transcriptome analyses were performed to investigate the evolution of the genus Acrostichum (Zhang et al. Citation2016). However, lack of the chloroplast genome sequence resulted in limited understanding of these mentioned data. Therefore, we reported the chloroplast genome of A. speciosum, which is a rare species in south China.
The plant material of A. speciosum was collected from the plantation of Dongzhaigang Mangrove Research Institute, Haikou, China (110°34′36.87″E, 19°57′6.77″N), and the voucher specimen is deposited in the herbarium of Hainan Normal University (specimen no. 19HNNU1223). The total genomic DNA was extracted from fresh leaves of A. speciosum by CTAB method (Doyle and Doyle Citation1987). The DNA was then randomly fragmented to 500 bp for library construction, which was sequenced PE-150 using Illumina Hiseq 2000 platform. A total of 5 Gb raw reads were obtained and assembled to generate the complete chloroplast genome using SOAPdenovo2 (Luo et al. Citation2012). Assembled chloroplast genome was annotated using online tool CPGAVAS (Shi et al. Citation2019).
The complete chloroplast genome of A. speciosum has a circular DNA of 156,095 bp with 38.45% of GC content (GenBank MT026711). A large single copy (LSC) of 84,476 and a small single copy (SSC) of 21,744 bp are separated by a pair of identical inverted repeat regions (IRs) of 24,938 bp each. Totally, 112 genes were annotated, including 72 protein-coding genes, 32 tRNAs, and 8 rRNAs.
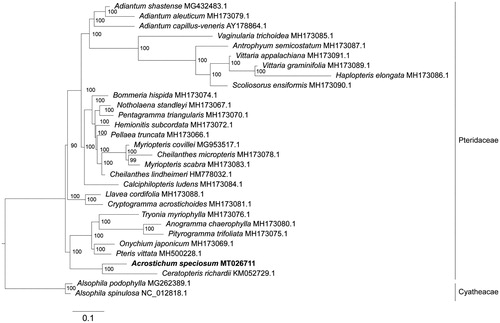
A total of 30 representative ferns were selected to construct a phylogenetic tree of Pteridaceae. Maximum-likelihood (ML) approach was performed using the complete chloroplast genome sequences of A. speciosum, 27 Pteridaceae species and two outgroups (Alsophila spinulosa and Alsophila podophylla). All sequences were aligned using MAFFT v7.455 (Katoh and Standley Citation2013). RAxML software (Stamatakis Citation2014) was used to build the phylogenetic tree with General Time Reversible + Gamma nucleotide substitution model + proportion of invariable sites (GTRGAMMAI) using 1000 bootstrap replicates. The result showed that A. speciosum and Ceratopteris richardii, both are aquatic plants, formed an original base clade in Pteridaceae, indicating that water ferns might have chloroplast genome sequence more similar to the ancestors (). This study provides a high-quality complete chloroplast genome of mangrove fern A. speciosum, which will be useful to better understanding the evolutionary history of ferns.
Figure 1. Phylogenetic analysis based on complete chloroplast genome sequences of 30 representative ferns, including A. speciosum, 27 species from Pteridaceae, and two outgroups from Alsophila genus. The phylogenetic tree was constructed by RAxML with bootstrap support values on each node (bootstrap was set to 1000). The GenBank accessions were indicated for each sequence.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18.
- Nabeelah BS, Fawzi MM, Gokhan Z, Rajesh J, Nadeem N, Rengasamy KKR, Albuquerque R, Pandian SK. 2019. Ethnopharmacology, phytochemistry, and global distribution of mangroves-A comprehensive review. Mar Drugs. 17:231.
- Robison TA, Grusz AL, Wolf PG, Mower JP, Fauskee BD, Sosa K, Schuettpelz E. 2018. Mobile elements shape plastome evolution in ferns. Genome Biol Evol. 10(10):2558–2571.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345. 31066451
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Yamamoto T, Tsuda Y, Mori GM, Cruz MV, Shinmura Y, Wee AK, Takayama K, Asakawa T, Yamakawa T, Suleiman M, et al. 2016. Development and characterization of 27 microsatellite markers for the mangrove fern, Acrostichum aureum (Pteridaceae). Appl Plant Sci. 4(9):apps.1600042.
- Zhang Z, He Z, Xu S, Li X, Guo W, Yang Y, Zhong C, Zhou R, Shi S. 2016. Transcriptome analyses provide insights into the phylogeny and adaptive evolution of the mangrove fern genus Acrostichum. Sci Rep. 6(1):35634.
- Zhang R, Liu T, Wu W, Li Y, Chao L, Huang L, Huang Y, Shi S, Zhou R. 2013. Molecular evidence for natural hybridization in the mangrove fern genus Acrostichum. BMC Plant Biol. 13(1):74.
