Abstract
Cuphea hyssopifolia is a small evergreen shrub of great economic and medicinal values. However, there are few studies on the ecological and genetic characteristics of this species. Chloroplast genomic analysis has been shown to resolve phylogenetic relationships. Here, we sequenced the complete chloroplast (cp) genome of C. hyssopifolia, which is the first complete cp genome reported in the genus Cuphea. The size of the whole cp genome is 158,821 bp, exhibiting the large (LSC, 89,064 bp) and small (SSC, 18,373 bp) single-copy regions and a pair of inverted repeats (IRs, 25,692 bp). The genome contained 111 unique genes, including 78 protein coding genes, 29 transfer RNAs (tRNA), and four ribosomal RNAs (rRNA). Neighbour-joining (NJ) phylogenetic analysis based on 43 genomes showed that C. hyssopifolia was closer to Heimia myrtifolia than Lagerstroemia species in the Lythraceae family.
Cuphea hyssopifolia is a small evergreen shrub of the genus Cuphea in the Lythraceae family. It is native to Mexico and Guatemala, and commonly cultivated in southern China (Gonzalez et al. Citation1994). As continuously flowers throughout the year, it has high ornamental value and is often used as a garden ornamental plant. Many horticultural varieties of C. hyssopifolia have been reported, such as Wescuflodia (Unger Citation2018) Magenta broder (Elliot Citation2011). It has also been reported to treat gastric diseases and cancers (Chen et al. Citation1999). In addition, the results of Lu et al showed that dense vegetation formed by C. hyssopifolia can limit the germination and growth of the invasive plant Macfadyena unguis-cati (Lu et al. Citation2005). Liu et al discovered that the extract of C. hyssopifolia has application potential in cosmetics and medical products (Liu et al. Citation2012). Therefore, the application prospect of C. hyssopifolia is very broad and is worthy of further research and development.
In 1986, the cp genome of Marchantia polymorpha was first sequenced (Umesono et al. Citation1984). To date, more than 4000 complete cp genome sequences have been stored in NCBI database. Chloroplasts, like mitochondria, is one of plastids. Because chloroplasts have maternal inheritance, they have become a research hots pot in recent years (Daniell et al. Citation2016). Little research has been done on the cp genome of C. hyssopifolia, so we assembled and characterized the complete cp genome of C. hyssopifolia with next-generation sequencing technology (GenBank: MN833211).
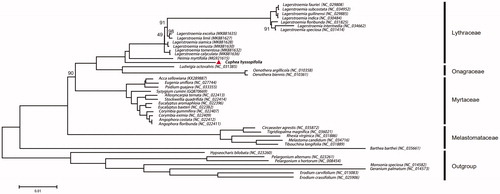
Fresh leaves were collected from the C. hyssopifolia grown in Zhejiang A & F University (30°26′N, 119°72′E) and immediately dried in silica gel. The voucher specimen is deposited at the Herbarium of Zhejiang A & F University (specimen code ZAFU2019066). CpDNA was then isolated following the CTAB protocol (Doyle and Doyle Citation1987). The 150-bp paired-end (PE) reads were generated by an Illumina HiSeq 2000 sequencer (Gu et al. Citation2019). Platanus v2.0 with default settings were employed to assemble the reads from cp genome of C. hyssopifolia. DOGMA v1.2 software was used to annotate the cp genes automatically (Wyman et al. Citation2004). The genome map of C. hyssopifolia was then drawn using OGDRAW (Lohse et al. Citation2013). We used MEGA 6 to generate a neighbour-joining tree () based on 64 homologous protein coding genes from 36 Myrtales species representing four families (Lythraceae, Myrtaceae, Onagraceae and Melastomataceae) (Tamura et al. Citation2013). To produce a rooted tree, we chose seven Geraniaceae species as outgroup.
Figure 1. The neighbour-joining tree based on 64 shared protein-coding genes of 36 Myrtales species and 7 outgroup species. Numbers near the branch mean the bootstrap value of the protein analysis for each clade, and the values equal to 100 are not displayed.
The structure of the C. hyssopifolia cp genome is typically quadripartite. A pair of 25,692 bp IRs divide the cp genome into a large single copy (LSC, 89,064 bp) and a small single copy (SSC, 18,373 bp) . The whole cp genome was 158,821 bp in length and the total GC content was 36.97%. The GC content of the LSC and SSC regions were 34.91 and 31.05%, respectively. The molecule annotation resulted in the identification of 111 unique genes, including 29 tRNAs, four rRNAs and 78 protein-coding genes. 7 tRNA genes, four rRNA genes and 6 coding genes are located within the IR regions. Of the 16 intron-containing genes, 13 genes (trnK, rps16, atpF, rpoC1, trnL-UAA, trnV-UAC, petB, petD, rpl16, ndhB, trnA, trnI-GAU, ndhA) have a single intron and three genes (clpP, rps12 and ycf3) have two introns.
In order to reveal the phylogenetic relationship between C. hyssopifolia and other species in Lythraceae, we selected 36 published cp genomes of Myrtales including 18 Lythraceae species, and seven Geraniaceae species as outgroup. The neighbour-joining phylogenetic tree based on the 43 cp genomes () showed that C. hyssopifolia was most closely related to Heimia myrtifolia in the Lythraceae with strong support. The complete cp genome presented in this study will provide useful data for population genomic and evolutionary studies on C. hyssopifolia.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen LG, Yen KY, Yang LL, Hatano T, Okuda T, Yoshida T. 1999. Macrocyclic ellagitannin dimers, cuphiin D1 and D2 and accompanying tannins from Cuphea hyssopifolia. Phytochemistry. 50(2):307–312.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Elliot R. 2011. Cuphea plant named ‘Magenta Border’: United States, USPP22157P2. 2011-09-20.
- Gonzalez AG, Valencia E, Exposito TS, Barrera JB, Gupta MP. 1994. Chemical components of Cuphea species. Carthagenol: a new triterpene from Cuphea carthagenesis. Planta Med. 60:592–593.
- Gu CH, Ma L, Wu ZQ, Chen K, Wang YX. 2019. Comparative analyses of chloroplast genomes from 22 Lythraceae species: inferences for phylogenetic relationships and genome evolution within Myrtales. BMC Plant Biol. 19(1):281–299.
- Liu JK, Ding ZH, Bai X, Jia RR, Qu Y, Dong ZJ. 2012. The application of the extract of Cuphea hyssopifolia in cosmetics and medical products:China, CN102525860A [P]. 2012-07-04.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41:575–581.
- Lu CY, Hu HY, Zhang MQ, Zhong YT, Zheng FZ. 2005. Studies on the control of alien invasive plant Macfadyena unguis-cati with ecological substitution. Plant Protection. 31(3):53–56.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Umesono K, Inokuchi H, Ohyama K, Ozeki H. 1984. Nucleotide sequence of Marchantia polymorpha chloroplast DNA: a region possibly encoding three tRNAs and three proteins including a homologue of E. coli ribosomal protein S14. Nucl Acids Res. 12(24):9551–9565.
- Unger H. 2018. Cuphea plant named ‘Wescuflodia’: United States, US20180228071P1 [P]. 2018-08-09.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
