Abstract
Ulva meridionalis is one of the causal green macroalgae for green tides in Japan and exists in coastal areas of China. In the present study, complete chloroplast (cp) genome sequence of Ulva meridionalis was reported, and the total length of this species was 88,653 bp (GenBank accession number MN889540). The overall base composition of cp genome was A (37.1%), T (39.0%), C (11.4%) and G (12.5%), similar to other Ulva macroalgae within cp genome, and the percentage of A + T (76.1%) was higher than C + G (23.9%). U. meridionalis cp genome encoded 113 genes, including 80 protein-coding genes, 27 transfer RNAs genes and 6 ribosomal RNAs genes. The maximum likelihood phylogenetic analysis shows that Ulva linza is the closest sister species of U. meridionalis.
Ulva species widely exist in the world, and there are more than 80 species of Ulva counted (Blomster et al. Citation1998; Guiry and Nic Citation2003; Hayden et al. Citation2003). On the one hand, Ulva species are important economic macroalgae (Vesty et al. Citation2015), such as Ulva linza and Ulva fasciata (Sivaprakash et al. Citation2019), which have the potential development value. On the other hand, the proliferation of Ulva species may cause green tides, such as Ulva prolifera, which causes challenges to the prevention of ecological disasters around the world (Huo et al. Citation2014; Zhang et al. Citation2014; Zhang, Shi, et al. Citation2019; Liu et al. Citation2019). China has the high frequency outbreak of green tides (Smetacek and Zingone Citation2013; Wang et al. Citation2018). In fact, green tides have erupted in the East Sea, Yellow Sea and Bohai Sea of China over the past years (Zhang et al. Citation2015; Cai et al. Citation2018; Zhao et al. Citation2019; Song et al. Citation2019), resulting in serious damage to the marine environment (Zhang, He, et al. Citation2019).
Ulva meridionalis is a marine tropical macroalgae that mainly distributed in southern Japan (Shimada et al. Citation2008). This species has not been collected from the Yellow Sea before. However, in August 2019, we found that U. meridionalis widely distributed in crab aquaculture ponds near the estuary of Qingdao, China (36°12′26.316″N, 120°06′37.756″E), and the specimen was stored in the herbarium of Shanghai Ocean University Museum (SHOU2019QD082001). According to reported researches, this species has a strong ecological adaptability, the maximum growth rate can reach up to 0.796 d−1 under the conditions that organic phosphorus and nitrogen concentrations are 0.125 mg/L and 4.02 mg/L (Yang Citation2018; Kang et al. Citation2020), which has a high risk to become new green tides in China. So it is of great significance to sequence the chloroplast (cp) genome of U. meridionalis for further research of evolutionary relationships. Here, we reported the complete cp genome of U. meridionalis first time, in order to effectively record genetic resources of this species and provide references for subsequent researches. Additionally, the phylogenetic status of U. meridionalis was analyzed.
We sent the specimen to Sangon Biotech (Shanghai) Co., Ltd. for high-throughput sequencing. The genomic shotgun library was prepared by using the TruSeq DNA Sample Prep Kit (Illumina, USA), paired-end sequences were obtained through the Illumina HiSeq 2500 platform later. Ulva flexuosa (Cai et al. Citation2017) and U. prolifera (Jiang et al. Citation2019) were taken as models for sequence splicing about the cp genome of U. meridionalis. Complete cp genome of U. meridionalis was 88,653 bp in length and was annotated in GenBank with the accession number MN889540. The overall base composition of cp genome was A (37.1%), T (39.0%), C (11.4%), G (12.5%), similar to other Ulva macroalgae in cp genome, and the percentage of A + T (76.1%) was higher than C + G (23.9%). The U. meridionalis cp genome encoded 113 genes, including 80 protein-coding genes, 27 transfer RNAs genes and 6 ribosomal RNAs genes.
Our laboratory had studied the cp genome of U. flexuosa (NC035823, KX579943), U. linza (NC030312; KX058323) (Wang et al. Citation2017) and U. prolifera (NC036137, KX342867) before. In addition, we downloaded sequences from the NCBI database: Ulva mutabilis (MK069584, NC043860), Ulva ohnoi (AP018696), Ulva sp. (KP720616), Ulva fasciata (KT882614, NC029040), Ulva lactuca (MH730972, NC042255), Ulva pertusa (MN853875) and Ulva compressa (MK069585). A Maximum-likelihood (ML) phylogenetic tree with 17 complete cp genome of Ulva and 1 outgroup called Pseudendoclonium akinetum (NC008114) was constructed by using the MEGA 7 software () (Kumar et al. Citation2016), which showed U. meridionalis is closely related to U. linza.
Figure 1. Maximum likelihood phylogenetic tree for U. meridionalis based on the chloroplast genomes. Numbers above each node indicate the bootstrap support value.
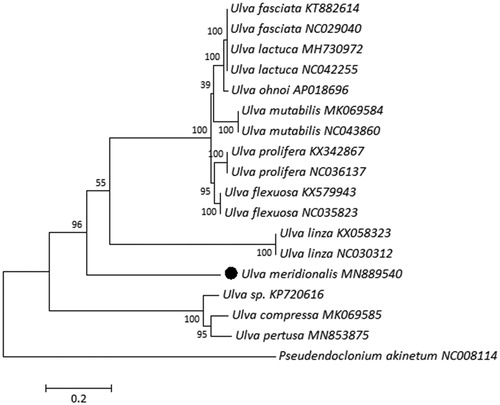
In this thesis, we reported a complete cp genome of U. meridionalis and provided detailed genetic information about this species, which will be useful for studying the genetic diversity and phylogenetic history of U. meridionalis and its related species.
Acknowledgments
We thank North China Sea Environmental Monitoring Center for providing personnel assistance and vehicle transportation service.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Blomster J, Maggs C, Stanhope M. 1998. Molecular and morphological analysis of Enteromorpha Intestinalis and E. compressa (Chlorophyta) in the British isles. J Phycol. 34(2):319–340.
- Cai CE, Wang LK, Jiang T, Zhou LJ, He PM, Jiao BH. 2018. The complete mitochondrial genomes of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta). Conserv Genet Resour. 10(3):415–418.
- Cai CE, Wang LK, Zhou LJ, He PM, Jiao BH. 2017. Complete chloroplast genome of green tide algae Ulva flexuosa (Ulvophyceae, Chlorophyta) with comparative analysis. PLoS ONE. 12(9):e0184196.
- Guiry MD, Nic DE. 2003. 56 Algaebase. J Phycol. 39(S1):19–20.
- Hayden H, Blomster J, Maggs C, Silva P, Stanhope M, Waaland J. 2003. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur J Phycol. 38(3):277–294.
- Huo YZ, Hua L, Wu HL, Zhang JH, Cui JJ, Huang XW, Yu KF, Shi HH, He PM, Ding DW. 2014. Abundance and distribution of Ulva microscopic propagules associated with a green tide in the southern coast of the Yellow Sea. Harmful Algae. 39:357–364.
- Jiang T, Gu K, Wang LK, Liu Q, Shi JT, Liu MM, Tang CY, Su YZ, Zhong SC, Cai CE, et al. 2019. Complete chloroplast genome of Ulva prolifera, the dominant species of green macroalgal blooms in Yellow Sea, China. Mitochondrial DNA Part B. 4(1):1930–1931.
- Kang XY, Liu JL, Yang XQ, Cui JJ, Zhao LJ, Wen QL, Fu ML, Zhang JH, He PM. 2020. The complete mitochondrial genome of a green macroalgae species: Ulva meridionalis (Ulvales: Ulvaceae). Mitochondrial DNA Part B. 5(1):760–761.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu JL, Zhao XH, Kang XY, Zhuang MM, Ding XW, Zhao LJ, Wen QL, Zhu Y, Gu K, Bao QJ, et al. 2019. Good news: we can identify Ulva species erupted in the Yellow Sea more easily and cheaply now. Conserv Genet Resour. Early Online, doi:10.1007/s12686-019-01114-x.
- Shimada S, Yokoyama N, Arai S, Hiraoka M. 2008. Phylogeography of the genus Ulva (Ulvophyceae, Chlorophyta), with special reference to the Japanese freshwater and brackish taxa. J Appl Phycol. 20(5):979–989.
- Sivaprakash G, Mohanrasu K, Ananthi V, Jothibasu M, Nguyen DD, Ravindran B, Chang SW, Phuong NT, Tran NH, Sudhakar M, et al. 2019. Biodiesel production from Ulva linza, Ulva tubulosa, Ulva fasciata, Ulva rigida, Ulva reticulate by using Mn2ZnO4 heterogenous nanocatalysts. Fuel. 255:115744.
- Smetacek V, Zingone A. 2013. Green and golden seaweed tides on the rise. Nature. 504(7478):84–88.
- Song W, Wang ZL, Li Y, Han HB, Zhang XL. 2019. Tracking the original source of the green tides in the Bohai Sea, China. Estuar Coast Shelf Sci. 219:354–362.
- Vesty EF, Kessler RW, Wichard T, Coates JC. 2015. Regulation of gametogenesis and zoosporogenesis in Ulva linza (Chlorophyta): comparison with Ulva mutabilis and potential for laboratory culture. Front Plant Sci. 6:15.
- Wang LK, Cai CE, Zhou LJ, He PM, Jiao BH. 2017. The complete chloroplast genome sequence of Ulva linza. Conserv Genet Resour. 9(3):1–4.
- Wang S, Huo Y, Zhang J, Cui J, Wang Y, Yang L, Zhou Q, Lu Y, Yu K, He P. 2018. Variations of dominant free-floating Ulva species in the source area for the worlds largest macroalgal blooms, China: Differences of ecological tolerance. Harmful Algae. 74:58–66.
- Yang Y. 2018. Study on nutrient removal from secondary effluent and water purification using Ulva Meridionalis. Xi’an: Shaanxi University of Science and Technology; p. 1–60.
- Zhang JH, Huo YZ, Wu HL, Yu KF, Kim JK, Yarish C, Qin YT, Liu CC, Xu R, He P. 2014. The origin of the Ulva macroalgal blooms in the Yellow Sea in 2013. Mar Pollut Bull. 89(1–2):276–283.
- Zhang JH, Liu CC, Yang LL, Gao S, Ji X, Huo YZ, Yu KF, Xu R, He PM. 2015. The source of the Ulva blooms in the East China Sea by the combination of morphological, molecular and numerical analysis. Estuar Coast Shelf Sci. 164:418–424.
- Zhang JH, Shi JT, Gao S, Huo YZ, Cui JJ, Shen H, Liu GY, He PM. 2019. Annual patterns of macroalgal blooms in the Yellow Sea during 2007-2017. Plos One. 14(1):e0210460.
- Zhang YY, He PM, Li HM, Li G, Liu JH, Jiao FL, Zhang JH, Huo YZ, Shi XY, Su RG, et al. 2019. Ulva prolifera green-tide outbreaks and their environmental impact in the Yellow Sea, China. Natl Sci Rev. 6(4):825–838.
- Zhao XH, Cui JJ, Zhang JH, Shi JT, Kang XY, Liu JL, Wen QL, He PM. 2019. Reproductive strategy of the floating alga Ulva prolifera in blooms in the Yellow Sea based on a combination of zoid and chromosome analysis. Mar Pollut Bull. 146:584–590.
