Abstract
The complete mitogenome of Gunungidia aurantiifasciata (GenBank accession number MT012904) is 15,472 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes, and a putative control region. The overall base composition is A (40.01%), C (11.93%), G (10.63%) and T (37.43%), demonstrating an obvious bias of high AT content (77.44%). ATG, ATA, ATT, TTG were initiation codons and TAA, TAG, and T were termination codons. All 22 tRNAs have the typical cloverleaf secondary structure, with an exception for trnS1 (AGN). The phylogenetic relationship based on neighbor-joining method showed that G. aurantiifasciata is clustered with two Typhlocybinae species, which agree with the conventional taxonomy.
The leafhopper Gunungidia aurantiifasciata belongs to the subfamily Typhlocybinae, which is the second largest leafhopper subfamily and includes about 5000 described species around the world (Dietrich and Dmitriev Citation2006). Most of these species caused significant losses in the cultivation of woody and herbaceous plants through feeding and virus transmission (Wang et al. Citation2017; Tan et al. Citation2020). In this study, adult species of G. aurantiifasciata were collected from Jianhe County, Guizhou Province, China (N26°38′, E108°28′), and deposited in the insect specimen room of Kaili University with an accession number KLU-Col-2019002.
The complete mitogenome of G. aurantiifasciata (GenBank accession number MT012904) is 15,472 bp in length, and contains 13 protein-coding genes (PCGs), 22 transfer RNA genes (tRNAs), two ribosomal RNA genes (16S rRNA and 12S rRNA), and a putative control region (Boore Citation1999). The gene order and organization of G. aurantiifasciata were identical with other Typhlocybinae species (Tan et al. Citation2020; Yuan et al. Citation2020). The overall base composition of G. aurantiifasciata mitogenome were A (40.01%), C (11.93%), G (10.63%) and T (37.43%), with an A + T bias (77.44%). This mitogenome appeared a positive AT-skew (0.033) and a negative GC-skew (−0.057). Fourteen genes (trnQ, trnC, trnY, trnF, nad5, trnH, nad4, nad4L, trnP, nad1, trnL1, trnV, 16S rRNA, and 12S rRNA) were transcribed on the minority strand (N-strand), whereas the others were encoded on the majority strand (J-strand).
A total of 48 bp overlaps had been found at 12 gene junctions and the longest overlap located between trnS2 and nad1. There are 13 intergenic spacer sequences in a total of 59 bp with length varying from 1 to 10 bp. The largest intergenic spacer is located between trnM and trnQ. The length of 22 tRNAs ranged from 61 bp (trnC) to 71 bp (trnK), A + T content ranged from 65.22% (trnQ) to 87.30% (trnE). All of the 22 tRNAs were predicted to have typical cloverleaf secondary structures, except the gene trnS1 lacking the dihydrouridine arm. The 16S rRNA was 1,185 bp in length with an A + T content of 84.85% and located between trnL1 and trnV. The 12S rRNA was 734 bp in length with an A + T content of 80.93% and located between trnV and CR. The control region was 1139 bp in length with an A + T content of 88.50%, which was located between 12S rRNA and trnI.
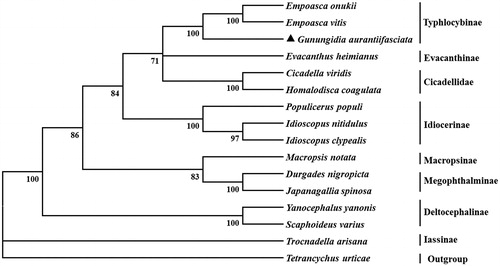
Twelve PCGs initiate with ATN as the start codons (ATG for atp6, cob, cox1, cox3, nad4, and nad4L; ATT for cox2 and nad6; ATA for atp8, nad1, nad2, and nad3), whereas nad5 use TTG as start codon. Eleven PCGs terminate with the conventional stop codon TAA, however, cob and cox2 use TAG and incomplete T as stop codon, respectively. Based on the concatenated amino aicd sequences of 13 PCGs, the neighbor-joining method was used to construct the phylogenetic relationship of G. aurantiifasciata and other 14 leafhoppers. The results showed that G. aurantiifasciata is clustered with two Typhlocybinae species, Empoasca onukii and Empoasca vitis (), which agree with the conventional taxonomy.
Figure 1. Phylogenetic tree showing the relationship between Gunungidia aurantiifasciata and 14 other leafhoppers based on neighbor-joining method. GenBank accession numbers used in the study are the following: Cicadella viridis (MK335936), Durgades nigropicta (KY123686), Empoasca onukii (MG190360), Empoasca vitis (KJ815009), Evacanthus heimianus (MG813486), Gunungidia aurantiifasciata (MT012904), Homalodisca coagulata (AY875213), Idioscopus clypealis (MF784430), Idioscopus nitidulus (KR024406), Japanagallia spinosa (NC035685), Macropsis notata (NC042723), Macrosteles quadrimaculatus (MG727894), Populicerus populi (MH492318), Scaphoideus varius (KY817245), Tetrancychus urticae (EU345430), Trocnadella arisana (NC036480), Yanocephalus yanonis (KY039113). T. urticae was used as an outgroup. Leafhopper determined in this study was marked with a triangle.
Disclosure statement
No potential conflict of interest was reported by the author(s). The authors also are responsible for the content and writing of the paper.
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Dietrich CH, Dmitriev DA. 2006. Review of the new world genera of the leafhopper tribe Erythroneurini (Hemiptera: Cicadellidae: Typhlocybinae). B Illinois Nat Hist Surv. 37:119–190.
- Tan C, Chen XX, Li C, Song YH. 2020. The complete mitochondrial genome of Empoascanara sipra (Hemiptera: Cicadellidae: Typhlocybinae) with phylogenetic consideration. Mitochondr DNA B. 5(1):260–261.
- Wang JJ, Li H, Dai RH. 2017. Complete mitochondrial genome of Taharana fasciana (Insecta, Hemiptera: Cicadellidae) and comparison with other Cicadellidae insects. Genetica. 145(6):593–602.
- Yuan XW, Li C, Song YH. 2020. Characterization of the complete mitochondrial genome of Mitjaevia protuberanta (Hemiptera: Cicadellidae: Typhlocybinae). Mitochondr DNA B. 5(1):601–602.
