Abstract
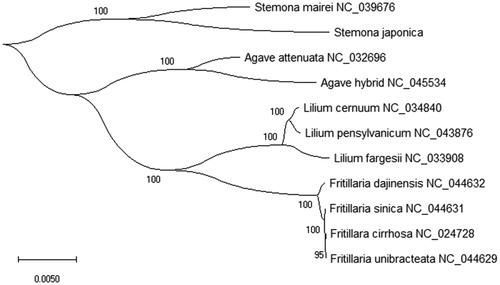
Stemona japonica is a species of perennial, flowering and herbaceous vines in the genus Stemona and the family Stemonaceae. In this study, we presented and annotated the complete chloroplast genome of S. japonica, which the chloroplast genome length was 154,224 bp, containing a large single-copy region (LSC) of 82,118 bp, a small single-copy region (SSC) of 17,938 bp and two same inverted repeat regions (IR) of 27,084 bp. The cp genome of S. japonica comprised 134 genes that included 88 protein-coding genes (PCGs), 38 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. The overall nucleotide of cp genome composition is: A of 47,338 bp (30.7%), T of 48,206 bp (31.3%), C of 29,528 bp (19.1%), and G of 29,101 bp (18.9%), with a total A + T content of 62.0% and G + C content of 38.0%. Phylogenetic Neighbor-Joining (NJ) analysis based on 11 plants species chloroplast genomes with Stemona japonica, the result shown that Stemona japonica is closely related to Stemona mairei in evolutionary relationship.
Stemona japonica is important one of herb plant in China, which belongs to the genus Stemona the family Stemonaceae (Lu et al. Citation2018). It is native to China, the Indian Subcontinent, Southeast Asia and northern Australia (Yang et al. Citation2006). In China, S. japonica is used as antitussive, expectorant and tonic in traditional Chinese medicine (CTM) and widely adopted in treating different diseases, which is also an important herb for the treatment of bone tuberculosis. Now, our understanding of the chloroplast genome of the S. japonica and other genome information is limited. In order to study the family Stemonaceae medicinal value of traditional Chinese medicine and natural product ingredients in genomics and bioinformatics, we presented and annotated the complete chloroplast genome of S. japonica and studied the phylogenetic relationship with other plants, which contributes to be useful development and utilization of traditional Chinese medicine.
The specimen sample of Stemona japonica was deposited from herb market near Zhejiang Chinese Medical University (Hangzhou, Zhejiang, China, 119.89E, 30.09 N). The chloroplast genome DNA was extracted from the root tissue using Plant Tissues Genomic DNA Extraction Kit (Solarbio, BJ, CN) and stored at Zhejiang Chinese Medical University (No. ZJCMU-003). The chloroplast genome DNA was sequenced by the Illumina HiSeq X Ten Sequencing Platform (Illumina Co., San Diego, CA). Quality control was performed by the FastQC (Andrews Citation2015) to remove low-quality reads and adapters. The chloroplast genome was assembled using the Plann (Huang and Cronk Citation2015) and annotated using the Geneious (Matthew et al. Citation2012). The physical map of the chloroplast genome of S. japonica was generated by the OGDRAW (Greiner et al. Citation2019), which was submitted to NCBI GenBank with accession No. MK9396752.
The complete chloroplast genome of Stemona japonica as a circle was 154,224 bp in length, which contains a large single-copy region (LSC) of 82,118 bp, a small single- copy region (SSC) of 17,938 bp and two inverted repeat regions (IRs) of 27,084 bp. The cp genome of S. japonica comprised 134 genes that included 88 protein-coding genes (PCGs), 38 transfer RNA (tRNA) genes and 8 ribosomal RNA (rRNA) genes. In one of IR region, 21 genes were found duplicated, which included 9 PCG genes species, 8 tRNA genes species and 4 rRNA genes species. The overall nucleotide of cp genome composition is: A of 47,338 bp (30.7%), T of 48,206 bp (31.3%), C of 29,528 bp (19.1%), and G of 29,101 bp (18.9%), with a total AT content of 62.0% and GC content of 38.0%.
For phylogenetic analysis, we selected other 10 plant species chloroplast genomes to study the relationship with Stemona japonica by the Neighbor-Joining (NJ) method. The phylogenetic NJ tree was performed and reconstructed using the MEGA X software with 5000 bootstrap values replicate (Kumar et al. Citation2018). All of the nodes were inferred with strong support by the NJ methods. The final tree was edited using the iTOL Web software (Letunic and Bork Citation2016). The reconstructed phylogenetic NJ tree showed that Stemona japonica was closely related to Stemona mairei (NC_039676) in the evolutionary relationship (). So, that study is very important for the evolutionary research of the family Stemonaceae, also can be useful development and utilization of traditional Chinese medicine.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucl Acids Res. 47(W1):W59–W64.
- Huang DI, Cronk QCB. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucl Acids Res. 44(W1):W242–W245.
- Lu QX, Ye WQ, Lu RS, Xu WQ, Qiu YX. 2018. Phylogenomic and comparative analyses of complete plastomes of croomia and stemona (Stemonaceae). IJMS. 19(8):2383.
- Matthew K, Richard M, Amy W, Steven S-H, Matthew C, Shane S, Simon B, Alex C, Sidney M, Chris D, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Yang XZ, Tang CP, Ke CQ, Ye Y. 2006. Chemical constituents of Stemona japonica. J Asian Nat Prod Res. 8(1–2):47–53.