Abstract
The first complete chloroplast (cp) genome sequences of Pittosporum tobira were reported in this study. The cp genome of P. tobira was 153,754 bp long, which included a large single copy (LSC) region of 85,010 bp and a small single copy (SSC) region of 18,740 bp separated by two inverted repeat (IR) regions of 25,002 bp. The cp genome of this species contained 113 unique genes, including 79 protein-coding genes, 30 transfer RNA genes and 4 ribosomal RNA genes. The overall GC content was 38.3%. Phylogenetic analysis of the complete cp genomes within the order Apiales suggested that P. tobira, the representative of Pittosporaceae, is sister to the families of Apiaceae and Araliaceae.
Pittosporum tobira (Thunb.) Aiton is an evergreen plant in the family Pittosporaceae. It is grown near the coasts in China and Japan and cultivated widely as an ornamental shrub in warm-temperate regions. Pittosporum tobira contains many bioactive substances, including phenolic acids, flavonoids, terpenoids, and other metabolites. These compounds are synthesized by different parts (leaf, root, seed, flower, and stem bark) of this species (Fan et al. Citation2011) and exert many biological effects including anti-thrombogenic, anti-microbial, anti-hemolytic, cytoprotective and antioxidant proprieties (Ogihara et al. Citation1989; Nickavar et al. Citation2004; Oh et al. Citation2014; El Dib et al. Citation2015; Rjeibi et al. Citation2017). To better understand the applications of P. tobira, the complete chloroplast (cp) genome of this species was sequenced and analyzed using high-throughput sequencing technology.
The specimen (lpssy0321) of P. tobira was collected from Liupanshui, Guizhou, China (N26°34′52″, E104°49′02″, 1,800 m) and was deposited in the herbarium of Liupanshui Normal University (LPSNU). The total genomic DNA was extracted and used for sequencing as previously described (Zhang et al. Citation2019). Approximately, 2 Gb raw data were used for de novo cp genome assembly with SPAdes (Bankevich et al. Citation2012) and all predicted genes were annotated using PGA (Qu et al. Citation2019). The complete cp genome sequences of P. tobira were deposited in GenBank database under the accession number MN968282.
The complete cp genome of P. tobira is 153,754 bp in length and shows the GC content of 38.3%. The cp genome displays a typical quadripartite structure, two copies of inverted repeats (IRs, 25,002 bp) separated by a large single-copy region (LSC, 85,010 bp) and a small single copy (SSC, 18,740 bp) region. A total of 113 unique genes were encoded, including 79 protein-coding genes (PCGs), 30 transfer RNA (tRNA) genes, and 4 ribosomal RNA (rRNA) genes. Of them, 6 PCGs (ndhB, rps12, rpl23, rps7, rpl2 and ycf2), 4 rRNAs (rrn16, rrn23, rrn4.5 and rrn5), and 7 tRNAs (trnA-UGC, trnl-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC) have two copies. Fourteen genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rpoC1, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA and trnV-UAC) contain one intron and three genes (clpP, rps12 and ycf3) have two introns.
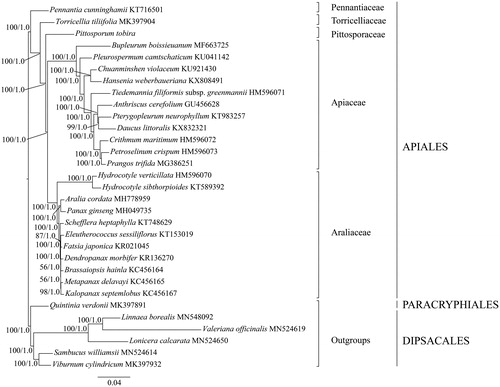
Pittosporum is the typical genus of the family Pittosporaceae. To confirm the phylogenetic position of Pittosporaceae, phylogenetic analysis was conducted within the order Apiales based on the maximum likelihood (ML) and Bayesian inference (BI) methods (Ronquist et al. Citation2012; Stamatakis Citation2014). The cp genomes of P. tobira and 24 species from other four families (Apiaceae (11), Araliaceae (11), Pennantiaceae (1), Torricelliaceae (1)) of Apiales were downloaded from GenBank. Five species (Linnaea borealis, Lonicera calcarata, Sambucus williamsii, Valeriana officinalis and Viburnum cylindricum) from Dipsacales and one species (Quintinia verdonii) from Paracryphiales were used as outgroups. The ML and BI analyses generated the same tree topology (). The resulted phylogenetic tree indicated that all species of Apiales were clustered into five distinct groups, corresponding to five distinct families (Apiaceae, Araliaceae, Pennantiaceae, Pittosporaceae, and Torricelliaceae) (). Pittosporum tobira, the representative of Pittosporaceae, is sister to the clade formed by Apiaceae and Araliaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- El Dib RA, Eskander J, Mohamed MA, Mohammed NM. 2015. Two new triterpenoid estersaponins and biological activities of Pittosporum tobira ‘Variegata’ (Thunb.) W. T. Aiton leaves. Fitoterapia. 106:272–279.
- Fan Y, Liu J, Wang Z. 2011. Advances in studies on chemical constituents from plants of Pittosporum Banks ex Gaertn. and their pharmacological activities. Chin Tradit Herb Drugs. 42(9):1842–1851.
- Nickavar B, Amin G, Yosefi M. 2004. Volatile constituents of the flower and fruit oils of Pittosporum tobira (Thunb.) Ait. grown in Iran. Z Naturforsch C J Biosci. 59(3–4):174–176.
- Ogihara K, Munesada K, Suga T. 1989. Sesquiterpene glycosides and other terpene constituents from the flowers of Pittosporum tobira. Phytochemistry. 28(11):3085–3091.
- Oh JH, Jeong YJ, Koo HJ, Park DW, Kang SC, Khoa HV, Le Le B, Cho JH, Lee JY. 2014. Antimicrobial activities against periodontopathic bacteria of Pittosporum tobira and its active compound. Molecules. 19(3):3607–3616.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):1–12.
- Rjeibi I, Ncib S, Ben Saad A, Souid S. 2017. Evaluation of nutritional values, phenolic profile, aroma compounds and biological properties of Pittosporum tobira seeds. Lipids Health Dis. 16(1):206.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Zhang SD, Zhang C, Ling LZ. 2019. The complete chloroplast genome of Rosa berberifolia. Mitochondr DNA B. 4(1):1741–1742.