Abstract
Lepus tibetanus pamirensis distributes only in the Pamir Plateau under a harsh environment with high attitude, cold and dry climates. The complete mitochondrial genome of the hare was determined (accession number MN539746) in a total length of 16,753 bp. The overall base composition of L. t. pamirensis mitogenome is 31.6% for A, 25.7% for C, 13.3% for G and 29.5% for T. It has the common characteristic with other mammals in regard to genome structure and gene arrangement. Based on published complete mitogenome sequences of lagomorphs, the phylogenetic tree suggested that L. t. pamirensis is closely related to L. yarkandensis and has far relationship with L. capensis. Our results would provide useful information for further study on conservation genetics and evolution of this hare species.
Lepus tibetanus pamirensis belongs to Lepus, Leporidae, Lagomorpha, Glires, Euarchontoglires and distributes only in the Pamir Plateau, a huge mountain junction crossed by five mountains in the center of Eurasia, which has typical continental plateau climate with cold, dry and strong solar radiation. Luo once assigned the hare as a subspecies of Lepus capensis–L. c. pamirensis according to morphological characteristics (Luo Citation1988). Wang (Citation2003) and Smith et al. (Citation2018) correct it as L. t. pamirensis. However, the genetic information about this hare under such extreme and isolated environment is still poorly understood. In this study, we sequenced the complete mitogenome of L. t. pamirensis and analyzed it.
The hare sample (TX-1) was collected in Tashkurgan Xinjiang in February 2018 (37°46′N, 75°13′E). After sampling, the specimen was stored in the animal specimen museum of Xinjiang Key Laboratory of Biological Resources and Genetic Engineering in Xinjiang University. The muscle tissue was used to extract total mitochondrial genomic DNA. Then the complete mitochondrial genome was sequenced by next-generation sequencing with Illumina Hiseq platform and was assembled with SOAPdenovo12.04 (http://soap.genomics.org.cn/soapdenovo.html.) (Luo et al. Citation2012). Genome components were analyzed by DOGMA2 (http://dogma.ccbb.utexas.edu/) and MITOS (http://mitos.bioinf.uni-leipzig.de/index.py). The phylogenetic tree was constructed using maximum likelihood (ML) method with MEGA7 software.
The complete mitogenome of L. t. pamirensis is 16,753 bp in length (GenBank accession number: MN539746) and contains all genes as in other mammalian mitogenomes (Shan and Liu Citation2016; Giannoulis et al. Citation2018; Li et al. Citation2018; Liu and Zhang Citation2018; Tan et al. Citation2018; Huang et al. Citation2019), including 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and a control region. The overall base composition of L. t. pamirensis mitogenome is 31.6% for A, 25.7% for C, 13.3% for G and 29.5% for T, displaying obvious anti-G bias and A + T-rich pattern (61.1%) of the vertebrate mitochondrial genomes. 14 of 22 tRNA genes are encoded on the heavy strand while the rest of 8 tRNAs are located on the light strand. 2 rRNA genes are both situated on the heavy strand. A majority of PCGs are encoded on the heavy strand with the exception of ND6 on the light strand.
Based on published 10 complete mitogenome sequences of Lepus from NCBI, and an Oryctolagus cuniculus as outgroup, phylogenetic analyses were performed with maximum likelihood (ML) method. Phylogenetic tree showed that L. t. pamirensis is closely related to the L. yarkandensis which is an endemic hare and also distributes in south of Xinjiang China (). Meanwhile, it displayed that Lepus tibetan pamirs has far relationship with L. capensis which comes from South Africa. The mitogenomic information in this study would contribute to further studies on molecular genetics, phylogeny and conservation of wildlife in L. t. pamirensis.
Figure 1. Phylogenetic analysis of 11 Lepus and an Oryctolagus cuniculus as outgroup based on the complete mitogenome using maximum likelihood method.
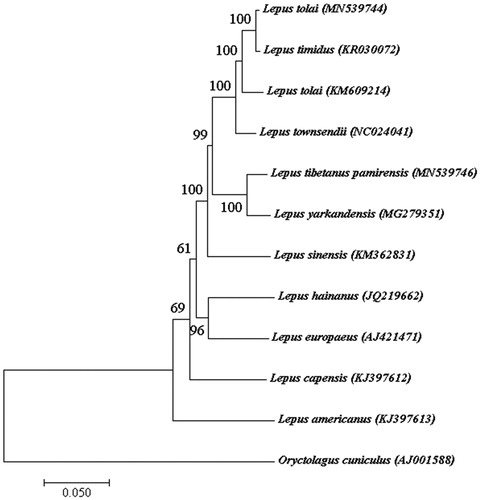
The sequence has been submitted to NCBI under the accession no. MN539744.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Giannoulis T, Stamatis C, Tsipourlianos A, Mamuris Z. 2018. Mitogenomic analysis in European brown hare (Lepus europaeus) proposes genetic and functional differentiation between the distinct lineages. Mitochondr DNA A. 29(3):353–360.
- Huang YL, Chen YX, Guo HT, Xu YH, Liu HY, Liu DW. 2019. The complete mitochondrial genome sequence of Yarkand hare (Lepus yarkandensis). Mitochondr DNA B. 4(2):3727–3728.
- Li FJ, Wang XM, Zhang Q, Jiang H, Wei HX, Wang Q, Chen SD, Liu SY. 2018. Complete mitochondrial genome of the Burmese Short-Tailed Shrew, Blarinella wardi, (Mammalia, Soricidae). Conservation Genet Resour. 10(2):233–236.
- Liu Y, Zhang M. 2018. The complete mitochondrial genome of Naemorhedus griseus, (Artiodactyla: Bovidae) and its phylogenetic implications. Conservation Genet Resour. 10(2):179–183.
- Luo ZX. 1988. The Chinese Hare. Beijing: China Forestry Publishing House.
- Luo RB, Liu BH, Xie YL, Li ZY, Huang WH, Yuan JY, He GZ, Chen YX, Pan Q, Liu YJ, et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaSci. 1(1):18–23.
- Shan WJ, Liu YG. 2016. The complete mitochondrial DNA sequence of the cape hare Lepus capensis pamirensis. Mitochondr DNA. 27(6):4572–4573.
- Smith AT, Johnston CH, Alves Pc, Hacklander K. 2018. Pikas, rabbits, and hares of the world. Baltimore: Johns Hopkins University Press.
- Tan ZD, Shen LY, Cheng X, Gan ML, Zhang SH, Zhu L. 2018. The complete mitochondrial genome sequence of Changbai Mountains wild boar (Cetartiodactyla: Suidae). Conservation Genet Resour. 10(1):99–102.
- Wang YX. 2003. A complete checklist of mammal species and subspecies in China–a taxonomic and geographic reference. Beijing: China Forestry Publishing House.
