Abstract
Oroxylum indicum (L.) Kurz is a medicinal plant commonly used in the southwest of China. In this study, we sequenced the complete chloroplast (cp) genome sequence of O. indicum to investigate its phylogenetic relationship in Bignoniaceae. The cp genome of O. indicum is 162,123 bp in length with 37.8% overall GC content, including a large single-copy (LSC) region of 86,416 bp and a small single-copy (SSC) region of 12,811 bp, which are separated by a pair of inverted repeats (IRs) of 31,448 bp. The cp genome contains 132 genes, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that O. indicum clustered together with Tr. Bignonieae Dumort.
Oroxylum indicum (L.) Kurz is the species of the genus Oroxylum within the family Bignoniaceae, which is a precious tree species in China, native in Guangxi, Fujian, Sichuan, Guizhou, and Yunnan (Peng et al. Citation2019). The seed of O. indicum is a traditional Chinese medicine that has been widely used for the treatment of cough, bronchitis, and other respiratory disorders for thousands of years (Dev et al. Citation2010). To date, there emerged many kinds of research on the pharmacological activity, chemical composition and quantitative analysis of O. indicum (Radhika et al. Citation2011). However, few data are available regardinggenomic studies on O. indicum.
Herein, we characterized the complete chloroplast (cp) genome of O. indicum. The GenBank accession number is MN933930. One O. indicum individual was collected from the medicinal plant arboretum, Yunnan Branch, Institute of Medicinal Plant, Chinese Academy of Medical Science (Yunan, China, 22°0′28″N, 100°47′17″E). The voucher specimen was deposited at the herbarium of Dali University, accession number: HBGP0595. Total DNA was isolated using the modified cetyl trimethyl ammonium bromide (CTAB) method (Doyle Citation1987). Paired-end reads were generated using Illumina NovaSeq system (Illumina, San Diego, CA, USA). In total, about 4.15 Gb of raw reads with 27,655,814 paired-end reads were obtained from high-throughput sequencing. Assembly and annotation were conducted by NOVOPlasty (Dierckxsens et al. Citation2017; Su et al. Citation2019) and GeSeq (Michael et al. Citation2017), respectively. The annotated cp genome was submitted to the GenBank under the accession number MN933930.
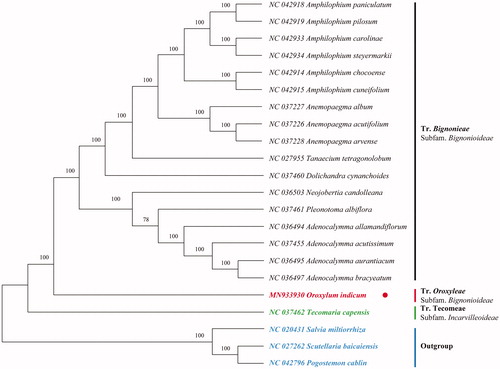
The complete cp genome of O. indicum is 162,123 bp in length, with a large single-copy (LSC) region of 86,416 bp, a small single-copy (SSC) region of 12,811 bp, and a pair of inverted repeats (IR) region of 31,448 bp. A total of 132 genes were annotated in this plastome, including 87 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The GC content of the whole cp genome is 37.8%. To reveal the phylogenetic position of O. indicum with other members of Bignoniaceae, a phylogenetic analysis was performed based on 18 complete cp genomes, and three taxa, Scutellaria baicalensis (NC_027262.1), Salvia miltiorrhiza (NC_020431.1) and Pogostemon cablin (NC_042796.1) were served as outgroups. The sequences were aligned by MAFFT v7.307 (Katoh and Standley Citation2013), and the maximum likelihood (ML) bootstrap analysis was conducted using RAxML (Stamatakis Citation2014); bootstrap probability values were calculated from 1000 replicates. The phylogenetic tree showed that O. indicum was closely related to tribe Bignonieae, and both tribes belonged to subfamily Bignonioideae (). In this paper, we firstly reported the complete cp genome of O. indicum, which will provide fundamental genetic resources for studying this important species as well as resolving its phylogenetic evolution.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dev LR, Anurag M, Rajiv G. 2010. Oroxylum indicum: a review. Pharmacogn J. 9(2):304–310.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18
- Doyle J. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1(19):11–15.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7 improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Michael T, Pascal L, Tommaso P, Elena SUJ, Axel F, Ralph B, Stephan G. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. W1(45):W6–W11.
- Peng Q, Shang XY, Zhu CC, Qin SY, Zhou Y, Liao QF, Zhang R, Zhao ZX, Zhang L. 2019. Qualitative and quantitative evaluation of Oroxylum indicum (L.) Kurz by HPLC and LC-qTOF-MS/MS. Biomed Chromatogr. 11(33):e4657.
- Radhika LG, Meena CV, Peter S, Rajesh KS, Rosamma MP. 2011. Phytochemical and antimicrobial study of Oroxylum indicum. Anc Sci Life. 30(4):114–120.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Su C, Liu PL, Chang ZY, Wen J. 2019. The complete chloroplast genome sequence of Oxytropis bicolor Bunge (Fabaceae). Mitochondrial DNA Part B. 4(2):3762–3763.