Abstract
Agrimonia pilosa Ldb., a herbaceous plant, has been used in Chinese medicines as anti-haemorrhagic, anthelmintic and anti-inflammatory agent. In this study, the complete chloroplast (cp) genome sequence of A. pilosa was first reported. The cp genome is 155,188 bp in length, with two inverted repeat (IR) regions of 25,965 bp, the large single copy (LSC) region of 84,521 bp and the small single copy (SSC) region of 18,737 bp. A total of 129 genes were predicted, including 84 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The phylogenetic analysis suggested that A. pilosa is more closely related to Sanguisorba filiformis with strong bootstrap values belonging to the subfamily Rosoideae.
Agrimonia pilosa Ldb. is a perennial herbaceous belonging to the Agrimonia genus in the Rosaceae family, which is widely distributed in Northern Asia and Eastern Europe (Seo et al. Citation2017). The whole plant are used as an anti-hemorrhagic, anthelmintic and anti-inflammatory agent in Chinese herbal medicine (Kasai et al. Citation1992). Previous studies on A. pilosa mainly focused on chemical components and medicinal activities (Park et al. Citation2019), and there was no research on complete cp genome sequence of A. pilosa. Here, we obtained the complete cp genome sequence of A. pilosa and revealed its phylogenetic relationships with other species in the Rosaceae, which would provide valuable genomic information to study the evolution of the genus Agrimonia.
Fresh leaves of A. pilosa were collected from Cangshan mountain (25°41′4″N, 100°08′31″E) of Dali county, Yunnan province, China, and the voucher specimen was deposited at the Herbarium of Dali University (HPGP0031). Total genomic DNA was extracted using the DNeasy plant mini kit (Qiagen) and subsequently sequence by Illumina NovaSeq system (Illumina, San Diego, CA, USA). Approximately, 4.09 Gb raw data (27,266,096 reads) was obtained from high-throughput sequence. The cp genome was assembled using NOVOPlasty v3.7 (Dierckxsens et al. Citation2017) and annotated with GeSeq (Michael et al. Citation2017). The annotated cp genome was submitted to the GenBank with an accession number of MT040192.
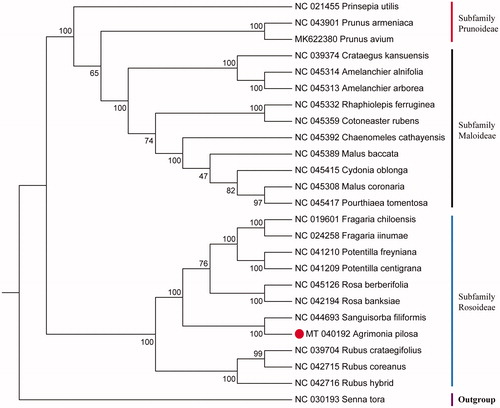
The complete cp genome of A. pilosa is 155,188 bp in length, with a typical quadripartite structure consisting of a large single copy (LSC) region (84,521 bp), a small single copy (SSC) region (18,737) and a pair of inverted repeat (IRs) regions (25,965 bp). The overall GC content of the cp genome is 36.91%. The plastid genome contained 129 genes, including 84 protein-coding genes, 8 ribosomal RNA genes, and 37 transfer RNA genes. To confirm the phylogenetic position of A. pilosa, 23 species of the Rosaceae family from NCBI were aligned using MAFFT v7.309 (Katoh and Standley, Citation2013). Senna tora (NC030193) was served as the out-group. The maximum likelihood (ML) bootstrap analysis was conducted using RAxML v8.2.12 (Stamatakis Citation2014), bootstrap probability values were calculated from 1000 replicates. The phylogenetic tree showed that A. pilosa was most closely related to Sanguisorba filiformis with strong bootstrap values belonging to the subfamily Rosoideae (). The chloroplast genome of A. pilosa and its phylogenetic analysis will provide useful information for phylogenetic and evolutionary studies in Rosoideae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kasai S, Watanabe S, Kawabata J, Tahara S, Mizutani J. 1992. Antimicrobial catechin derivatives of Agrimonia pilosa. Phytochemistry. 3(31):787–789.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 4(30):772–780.
- Michael T, Pascal L, Tommaso P, U E S, Axel F, Ralph B, Stephan G. 2017. GeSeq—versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Park J, Choi YG, Yun N, Xi H, Min J, Kim Y, Oh S. 2019. The complete chloroplast genome sequence of Viburnum erosum (Adoxaceae). Mitochondr DNA B. 2(4):3278–3279.
- Seo UM, Nguyen DH, Zhao BT, Min BS, Woo MH. 2017. Flavanonol glucosides from the aerial parts of Agrimonia pilosa Ledeb. and their acetylcholinesterase inhibitory effects. Carbohyd Res. 445:75–79.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 9(30):1312–1313.