Abstract
The shrews, Crocidura andamanensis, Crocidura nicobarica (order Eulipotyphla), and treeshrew, Tupaia nicobarica, (order Scandentia) are highly threatened mammals; endemic to Andaman and Nicobar archipelago. The aimed study provides the first baseline mitochondrial cytochrome b (mtCytb) genetic information of Crocidura and Tupaia species from Andaman and Nicobar Island. The mtCytb data were well discriminated against to all three studied species from their congeners with sufficient genetic distance. Crocidura andamanensis showed less genetic distance (13%) with Crocidura grayi, while high (21.3%) with Crocidura phuquocensis. Besides, the C. nicobarica showed less distance (5.5%) with Crocidura vosmaeri and Crocidura beccarii, while high (19%) with C. phuquocensis. Both C. andamanensis and C. nicobarica also depicted a 15.6% genetic distance with each other. Further, the treeshrew species T. nicobarica is apparently discriminated by other congeners with a sufficient genetic distance ranging from 8.8 to 25.5%. The maximum-likelihood (ML) topology distinctly discriminated against all the shrews and treeshrews species with significant bootstrap supports. The aimed study fortifies the efficacy of mtCytb gene to segregate the above discussed non-volant mammal species. We recommended the widespread noninvasive sampling and genotyping to elucidate the population structure, which assists to formulate the precise conservation measures to protect these unique mammals.
1. Introduction
The non-volant small mammals are generally known by rodents, shrews, treeshrews, hedgehog, and pika (Molur et al. Citation2005). They are widespread in all the continents except Antarctica (Wilson and Reeder Citation2005) and play a vital role in maintaining the healthy eco-system (Ryszkowski Citation1975). Among them, shrews help to increase soil fertility and control the population of invertebrates, while the treeshrew aids in seed dispersal in the tropical forest ecosystem. Based on the morphological characters, these two small mammals were grouped into two orders Eulipotyphla and Scandentia. The members of the order Eulipotyphla are commonly known as insectivorous, mouse-like mammals; and lives in sub-leaf stratum and burrow. However, the Scandentia (treeshrews) are primitive arboreal species and resembles with squirrel and mongoose (Li and Ni Citation2016). As of now, a total of 385 shrew species of 26 genera and 20 treeshrew species of five genera have been reported worldwide (Wilson and Reeder Citation2005). In India, 11 shrew species (white-toothed) and three treeshrew species were recorded to date (Molur et al. Citation2005). Among the extant Indian species, seven shrew species and two treeshrew species are distributed in the mainland, while the others are reported from islands (Molur et al. Citation2005). The unique tropical forest ecosystems of the Andaman and Nicobar group of Islands exhibit great biological diversity and endemism. So far, four shrew species (Crocidura andamanensis, Crocidura hispida, Crocidura jenkinsi, and Crocidura nicobarica) and one treeshrew species (Tupaia nicobarica) are restricted to Andaman and Nicobar archipelago. The survey on the status and distribution of these mammals in Andaman and Nicobar group of Islands was started long back and still continuing (Miller Citation1902; Ellerman and Morrison-Scott Citation1951; Saha Citation1980; Das Citation1999; Chakraborty et al. Citation2004; Hutterer Citation2005). Further, the ecology and behavior of these small mammals have been studied from Great Nicobar Island (Oommen and Shanker Citation2008). The recent study reported that the extant shrew and treeshrew species are threatened due to habitat loss, anthropogenic activities, forest fragmentation, tsunami events, and predation by other animals (Molur et al. Citation2005; IUCN Citation2019). However, the systematics studies for these group is poorly attempted due to the taxonomic uncertainties or challenging to access the specimens or biological samples (Hutterer Citation1993; Roberts et al. Citation2011). Hence, the scenario of phylogenetic evolution and biogeographic patterns of this group is still blurred to the scientific community.
In the early stage, the phylogenetic evolution and biogeography of these taxa were assessed by allozyme electrophoresis (Ruedi Citation1996). Further, with the advancement of molecular technique and its utility in systematics, the researchers have generated the genetic information of shrew and treeshrew species from different parts of the world to know their phylogeny and evolutionary pattern. The phylogeny, phylogeography, and geographical variation of shrew species, especially for the genus Crocidura were largely evaluated through mitochondrial cytochrome b (mtCytb) gene (Ruedi et al. Citation1998; Ohdachi et al. Citation2004; Querouil et al. Citation2011; Stanley et al. Citation2015) and multi-gene approaches (Nicolas et al. Citation2019). Further, the mitochondrial 12S and 16S rRNA genes were also used to understand the molecular phylogeny of treeshrews and their timescale of diversification (Olson et al. Citation2005; Roberts et al. Citation2011). Nevertheless, the genetic information of two highly threatened species (C. andamanensis and C. nicobarica) has never been assessed previously. Although the genetic information of T. nicobarica was generated from Little Nicobar Island and available in GenBank, the mtCytb genetic data has never been generated for the species from their type locality. Here, the aim of the present study is to generate the partial mtCytb data of two critically endangered shrews and one endangered treeshrew species from Andaman and Nicobar archipelago and compare them with the available database sequences to know their gene-specific clustering and genetic differences. The present genetic information would be helpful for population genetics of this threatened taxa, as well as facilitate to formulate the precise managements to conserve these small mammals.
2. Materials and methods
2.1. Survey and sampling
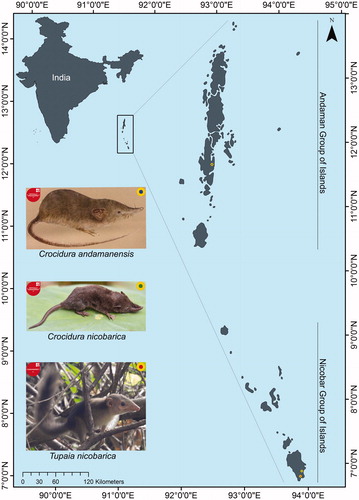
The recent fieldwork was conducted by the team of Zoological Survey of India (ZSI) during the ‘Great Nicobar expedition’ from November to December 2018. During the survey, a fresh natural kill of shrew specimen (ZSI_NC1) was spotted at Galathea, Great Nicobar Island (6.87N, 93.88E) (). Further, one morphological identified shrew, C. andamanensis (ZSI_NC2) and two subsamples (ZSI_NC3A, ZSI_NC3B) of one treeshrew, T. nicobarica were accessed from the National Zoological Collections (NZC) of Andaman and Nicobar Regional Center (ANRC), ZSI, Port Blair, India. No animals were killed, hence no prior permission was required in the present study. All biological samples were preserved in 70% ethanol in the Mammal and Osteology section, ZSI, Kolkata under voucher IDs (ZSI_NC1: Reg. No. 28533; ZSI_NC2: Reg. No. 28534; ZSI_NC3A: Reg. No. 28531; and ZSI_NC3B: Reg. No. 28532) for downstream molecular investigation.
Figure 1. Map showing the geographical locations of Andaman and Nicobar group of Islands in India and the collection locality of the endemic and threatened shrew and treeshrew species. The drawing of C. andamanensis was obtained from the published book of ZSI, MoEF&CC. The photograph of C. nicobarica was captured by the third author M.K., and T. nicobarica by G. Gokulakrishnan, ANRC, ZSI.
2.2. DNA extraction, PCR amplification, and sequencing
The genomic DNA was extracted by standard phenol-chloroform isoamyl alcohol protocol (Sambrook and Russell Citation2001) and checked through 1% agarose gel electrophoresis. The published primer pair for vertebrate taxa (mcb 398: 5′-TACCATGAGGACAAATATCATTCTG-3′ and mcb 869: 5′-CCTCCTAGTTTGTTAGGGATTGATCG-3′) (Verma and Singh Citation2002) was employed to amplify the partial mtCytb gene segment in a Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA) with the published thermal profile. The 25 µl PCR mixture comprises 10 pmol of each primer, 20 ng of DNA template, 1X PCR buffer, 1.0–1.5 mM of MgCl2, 0.25 mM of each dNTPs, and 1 U of Taq polymerase (Takara Bio Inc., Shiga, Japan). The amplified PCR products were checked in 1% agarose gel and purified by using a QIAquickR Gel extraction kit (QIAGEN Inc., Germantown, MD). The bi-directional sequencing of each sample was carried out by 48 capillary arrays 3730 DNA Analyzer (Applied Biosystems, Foster City, CA) available at ZSI, Kolkata, following the standard Sanger sequencing methods.
2.3. Sequence quality control, dataset preparation, and analysis
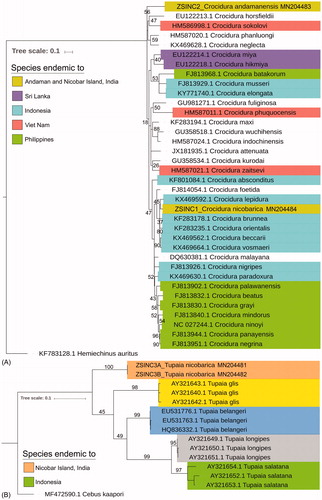
Both the generated forward and reverse chromatograms were checked by using SeqScanner Version 1.0 (Applied Biosystems Inc., Foster City, CA) and assembled by BioEdit v7.2.5 (Hall Citation1999). The consensus sequences were further checked through the online nucleotide BLAST program (https://blast.ncbi.nlm.nih.gov/), and ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to scrutinize and eliminate the insertion-deletion, and start-stop codons. The final sequences were submitted in the global GenBank database to acquire the accession numbers. To confirm the preliminary species identification, the generated sequences were checked in online identification systems in GenBank. Based on the similarity search result, 46 database sequences (34 sequences of shrew species from South and Southeast Asia +12 sequences of four treeshrew species) were acquired from GenBank and aligned by using ClustalX software (Thompson et al. Citation1997) to make a combined dataset. The genetic distance and topology were estimated through the Kimura 2 parameter (K2P) and maximum-likelihood (ML) tree through MEGAX (Kumar et al. Citation2018). The database sequences of a hedgehog, Hemiechinus auritus (KF783128), and a capuchin monkey, Cebus kaapori (MF472590) were used as an out-group in ML analysis.
3. Results and discussion
Island ecosystems act as a discrete biogeographic unit and, therefore, are believed to be a significant model for evolutionary studies. Although several island systems have been well explored and screened through genetic data to presume the biogeographic connectivity. Nevertheless, the odd distribution and ecology of any species induce uneven speciation in the Islands ecosystem, and extinction among groups (Upham et al. Citation2019). Further, the multi-locus phylogeny has been tested to clarify the diversification and evolution of Crocidura species from mainland of Africa (Nicolas et al. Citation2019), and the Island of Philippines and Taiwan (Esselstyn and Oliveros Citation2010; Giarla and Esselstyn Citation2015), Java (Esselstyn et al. Citation2013), Sundaland (Demos et al. Citation2016), as well as from Sulawesi (Eldridge et al. Citation2018). The genetic data is used to know the relationships between the higher taxonomic levels of Crocidura species (Dubey et al. Citation2007; He et al. Citation2010). Adding to this, the morphological character displacement is also evidenced in Crocidura species despite their asymmetric gene flow (Demos et al. Citation2016) and the geological and climatic changes to drive the speciation of the shrew in southeast Asian archipelagos (Esselstyn et al. Citation2009) is discussed. Besides, the molecular data also clearly supported the treeshrew topology under Scandentia (Schmitz et al. Citation2000). Nevertheless, the multi-gene assessment has been well assessed for treeshrews to know their phylogenetic relationship and evolution (Olson et al. Citation2005; Roberts et al. Citation2011). However, the Andaman and Nicobar Island in India is still believed to be unexplored and the genetic information of endemic shrews and treeshrew is still infancy.
Both the collected shrew specimens (ZSI_NC1 and ZSI_NC2) were identified as C. nicobarica and C. andamanensis by their external morphological characters as compared with the archival collections of ZSI and published information (Miller Citation1902). The identification was further confirmed based on their specific type locality collection, as C. nicobarica from Galathea, Great Nicobar Island, and C. andamanensis from Wrightmyo, South Andaman Island (11.79N, 92.71E) (). The identification of T. nicobarica (ZSI_NC3) was also confirmed by external morphology and type locality collection from Great Nicobar Island (6.83N, 93.87E). The generated mtCytb sequence of C. nicobarica (accession no. MN204484), and C. andamanensis (accession no. MN204483) showed 95.06% and 88.83% similarity with their nearest species Crocidura beccarii and Crocidura grayi, respectively, in GenBank database. The overall mean K2P genetic distance of the Crocidura shrew dataset was 15.2% and the ML tree distinctly discriminates all the Crocidura species (). The inter-species genetic distance of the studied shrew dataset was ranging from 1.8% (two Philippines species Crocidura negrina and Crocidura panayensis) to 21.5% (two Vietnamese species Crocidura phuquocensis and Crocidura sokolovi). The estimated genetic distance of C. nicobarica revealed less (5.5%) with two Indonesian species, Crocidura vosmaeri and Crocidura beccarii, while high (19%) with the Vietnamese species, Crocidura phuquocensis. Further, C. andamanensis showed less genetic distance (13%) with the Philippines endemic species, C. grayi; while high (21.3%) with C. phuquocensis. Moreover, the species endemic to specific regions showed high genetic distance, 15.6% between the two Indian species (C. nicobarica and C. andamanensis), 12.4% between the two Sri Lankan species (Crocidura miya and Crocidura hikmiya), 18.8% between three Vietnamese species (C. phuquocensis, C. sokolovi, and Crocidura zaitsevi), 10.1% between eight Philippines species, and 11.5% between 10 Indonesian species; which manifested the unparallel speciation within the South and Southeast Asian archipelago. The generated sequences of T. nicobarica (accession nos. MN204481 and MN204482) showed 85.68% similarity and 17.5% K2P genetic distance with the widely distributed northern treeshrew, Tupaia belangeri. The overall mean genetic distance of the present treeshrew dataset was 18.8%. All the five species (T. nicobarica, Tupaia glis, Tupaia longipes, Tupaia salatana, and T. belangeri) clearly discriminated by K2P genetic distances and ML clustering (). The highest intra-species genetic distance (4.2%) was observed in T. salatana, while the inter-species genetic distance was ranging from 8.8% (T. salatana and T. longipes) to 25.5% (T. salatana and T. glis). The aimed study provides the first baseline genetic data of endemic Crocidura species from Andaman and Nicobar Island. The study also provides the first mtCytb molecular data of T. nicobarica from Great Nicobar Island, which is thought to be a distinct subspecies. The generated sequences are enriched the global reference library and help to identify these species hereafter. The present effort also evidenced the efficacy of partial fragment of mtCytb gene to discriminate between the studied shrew and treeshrew species as compared to other South and Southeast Asian species. We assured that the extensive survey of these mammals from Andaman and Nicobar group of islands and their genetics study with multiple molecular markers would be helpful to clarify their population structure as well as formulate the accurate conservation measures to protect these endemic species.
Figure 2. The maximum-likelihood (ML) tree distinctly discriminated the studied shrew (A) and treeshrew species (B) with other South and Southeast Asian congeners. The database sequences of a hedgehog, H. auritus, and a capuchin monkey, C. kaapori were used to build the tree of Crocidura shrew and Tupaia treeshrew, respectively.
Author contributions
S.K., V.K., and K.C. conceived and designed the experiment. M.K., C.V., and C.S. collected the specimens, performed taxonomic identification, and captured photographs. K.C., V.K., and K.T. contributed to chemicals and laboratory facilities. K.T., and S.K. generated molecular data and data acquisition. S.K., K.T., and V.K. analyzed the data and prepared figures. S.K., K.T., and M.K. wrote the draft manuscript. All authors reviewed the manuscript.
Acknowledgments
The authors are thankful to the Director, Zoological Survey of India (ZSI), Ministry of Environment, Forests and Climate Change (MoEF&CC), Govt. of India for providing necessary facilities, constant support, and encouragement throughout the study. We also thank all team members of the Great Nicobar expedition for survey and sampling.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chakraborty S, Srinivasulu C, Srinivasulu B, Pradhan MS, Nameer PO. 2004. Checklist of insectivores (Mammalia: Insectivora) of South Asia. Zoos Print J. 19(2):1361–1371.
- Das I. 1999. A noteworthy collection of mammals from Mt Harriet, Andaman Islands, India. J South Asian Nat Hist. 4:181–185.
- Demos TC, Achmadi AS, Giarla TC, Handika H, Maharadatunkamsi , Rowe KC, Esselstyn JA. 2016. Local endemism and within island diversification of shrews illustrate the importance of speciation in building Sundaland mammal diversity. Mol Ecol. 25(20):5158–5173.
- Demos TC, Achmadi AS, Handika H, Maharadatunkamsi , Rowe KC, Esselstyn JA. 2016. A new species of shrew (Soricomorpha: Crocidura) from Java, Indonesia: possible character displacement despite interspecific gene flow. J Mammal. 98:183–193.
- Dubey S, Salamin N, Ohdachi SD, Barriere P, Vogel P. 2007. Molecular phylogenetics of shrews (Mammalia: Soricidae) reveal timing of transcontinental colonizations. Mol Phylogenet Evol. 44(1):126–137.
- Eldridge RA, Achmadi AS, Giarla TC, Rowe KC, Esselstyn JA. 2018. Geographic isolation and elevational gradients promote diversification in an endemic shrew on Sulawesi. Mol Phylogenet E. 118:306–317.
- Ellerman JR, Morrison-Scott T. 1951. Checklist of Palaearctic and Indian mammals 1758 to 1946. London (UK): British Museum (Natural History).
- Esselstyn JA, Maharadatunkamsi , Achmadi AS, Siler CD, Evans BJ. 2013. Carving out turf in a biodiversity hotspot: multiple, previously unrecognized shrew species co-occur on Java Island, Indonesia. Mol Ecol. 22:4972–4987.
- Esselstyn JA, Oliveros CH. 2010. Colonization of the Philippines from Taiwan: a multi-locus test of the biogeographic and phylogenetic relationships of isolated populations of shrews. J Biogeogr. 37:1504–1514.
- Esselstyn JA, Timm RM, Brown RM. 2009. Do geological or climatic processes drive speciation in dynamic archipelagos? The tempo and mode of diversification in Southeast Asian shrews. Evolution. 63(10):2595–2610.
- Giarla TC, Esselstyn JA. 2015. The challenges of resolving a rapid, recent radiation: empirical and simulated phylogenomics of Philippine shrews. Syst Biol. 64(5):727–740.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- He K, Li YJ, Brandley MC, Lin LK, Wang YX, Zhang YP, Jiang XL. 2010. A multi-locus phylogeny of Nectogalini shrews and influences of the paleoclimate on speciation and evolution. Mol Phylogenet Evol. 56(2):734–746.
- Hutterer R. 2005. Order Soricomorpha. In: Wilson DE and Reeder DM, editors. Mammal species of the world. Baltimore (MD): Johns Hopkins University Press; p. 220–311.
- Hutterer RM. 1993. Order Insectivora. In Wilson DE and Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference. Washington, DC: Smithsonian Institution Press; p. 69–130.
- IUCN. 2019. The IUCN red list of threatened species, Version 2019.1. Gland (Switzerland): IUCN; [Accessed 2019 July 16]. http://www.iucnredlist.org.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Li Q, Ni X. 2016. An early Oligocene fossil demonstrates treeshrews are slowly evolving “living fossils”. Sci Rep. 6(1):18627.
- Miller GS. 1902. Mammals of the Andaman and Nicobar Islands. Proc US Nat Mus. 24(1269):751–795.
- Molur S, Srinivasulu C, Srinivasulu B, Walker S, Nameer PO, Ravikumar L. 2005. Status of non-volant small mammals: Conservation Assessment and Management Plan (C.A.M.P) workshop report. Coimbatore (India): Zoo Outreach Organisation/CBSG-South Asia; p. 618.
- Nicolas V, Jacquet F, Hutterer R, Konečný A, Kouassi SK, Durnez L, Lalis A, Colyn M, Denys C. 2019. Multilocus phylogeny of the Crocidura poensis species complex (Mammalia, Eulipotyphla): influences of the palaeoclimate on its diversification and evolution. J Biogeogr. 46(5):871–883.
- Ohdachi SD, Iwasa MA, Nesterenko VA, Abe H, Masuda R, Haberl W. 2004. Molecular phylogenetics of Crocidura shrews (Insectivora) in east and central Asia. J Mammal. 85(3):396–403.
- Olson LE, Sargis EJ, Martin RD. 2005. Intraordinal phylogenetics of treeshrews (Mammalia: Scandentia) based on evidence from the mitochondrial 12S rRNA gene. Mol Phylogenet Evol. 35(3):656–673.
- Oommen MA, Shanker K. 2008. Ecology and behaviour of an endemic treeshrew Tupaia nicobarica Zelebor 1869 on Great Nicobar Island, India. J Bombay Nat Hist Soc. 105:55–63.
- Querouil S, Hutterer R, Barriere P, Colyn M, Peterhans JCK, Verheyen E. 2011. Phylogeny and evolution of African shrews (Mammalia: Soricidae) inferred from 16s rRNA sequences. Mol Phylogenet E. 20:185–195.
- Roberts TE, Lanier HC, Sargis EJ, Olson LE. 2011. Molecular phylogeny of treeshrews (Mammalia: Scandentia) and the timescale of diversification in Southeast Asia. Mol Phylogenet Evol. 60(3):358–372.
- Ruedi M. 1996. Phylogenetic evolution and biogeography of Southeast Asian shrews (genus Crocidura: Soricidae). Biol J Linn Soc. 58:197–219.
- Ruedi M, Auberson M, Savolainen V. 1998. Biogeography of Sulawesian Shrews: testing for their origin with a parametric bootstrap on molecular data. Mol Phylogenet Evol. 9(3):567–571.
- Ryszkowski L. 1975. The ecosystem role of small mammals. Ecol Bull. 19:139–145.
- Saha SS. 1980. Notes on some mammals recently collected from Andaman and Nicobar Islands. Rec Zool Surv Ind. 77:119–126.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press.
- Schmitz J, Ohme M, Zischler H. 2000. The complete mitochondrial genome of Tupaia belangeri and the phylogenetic affiliation of Scandentia to other eutherian orders. Mol Biol Evol. 17(9):1334–1343.
- Stanley WT, Hutterer R, Giarla TC, Esselstyn JA. 2015. Phylogeny, phylogeography and geographical variation in the Crocidura monax (Soricidae) species complex from the montane islands of Tanzania, with descriptions of three new species. Zool J Linn Soc. 174(1):185–215.
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25(24):4876–4882.
- Upham NS, Esselstyn JA, Jetz W. 2019. Ecological causes of uneven diversification and richness in the mammal tree of life. BioRxiv. DOI:10.1101/504803
- Verma SK, Singh L. 2002. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol Ecol Notes. 3(1):28–31.
- Wilson DE, Reeder DM, (eds.). 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd Ed. Baltimore (MD): Johns Hopkins University Press; p. 1–2141.
