Abstract
The Trispot Darter (Etheostoma trisella) is an imperiled small-bodied freshwater fish that inhabits headwaters of the upper Coosa River watershed in Alabama, Georgia, and Tennessee. We sequenced the complete mitochondrial genome of this species to develop non-invasive environmental DNA (eDNA) surveillance protocols. A mitochondrial phylogenomic analysis reveals the Trispot Darter to have diverged from soon after the common ancestor of genus Etheostoma. The mitochondrial genome sequence of E. trisella is similar to other darter species in terms of GC content and gene order, but the sequence is sufficiently divergent to permit the design of species-specific eDNA primers.
The Trispot Darter (Etheostoma trisella) is an imperiled, small-bodied (<6 cm) freshwater fish with unique life history traits (NatureServe Citation2013). Unlike most members of the subfamily Etheostominae, E. trisella is a migratory species that utilizes ephemeral streams as spawning habitat (Ryon Citation1986). Ephemeral streams are not afforded the same environmental protections as other freshwater habitats, and they are frequently disturbed by land-use changes. Etheostoma trisella is endemic to headwaters of the upper Coosa River watershed, where it is estimated that 80% of the species historical habitat has been lost to conversion to agriculture. In 2019, the species gained protection by the U.S. Endangered Species Act, and is currently listed as ‘Threatened’. Governmental agencies and non-governmental organizations recognize a need for periodic updates on habitat use and population status of E. trisella. Mitochondrial environmental DNA (eDNA) surveillance is affordable and noninvasive, and has demonstrated utility for monitoring imperiled freshwater fishes (Piggott Citation2016; Schroeter et al. Citation2019).
We sequenced the complete mitogenome of E. trisella to facilitate the design and development of eDNA detection protocols. A single specimen was collected from the Coosawattee River, Georgia (34.5731 N, −84.87556 W), in August 2018. A fin clip was taken following euthanasia, and the specimen was accessioned at the Georgia Department of Natural Resources (BA10-042). DNA was extracted using a Qiagen DNeasy kit. DNA quality and quantity were evaluated with a Qiaxcel and Nanodrop 2000 instruments, respectively. Shotgun library preparation and high-throughput sequencing were performed at the UC Berkeley DNA Sequencing Center.
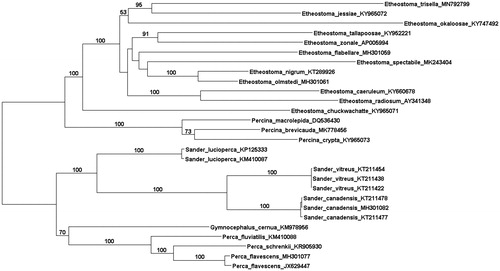
The sample was barcoded and sequenced on 2.9% of a pooled single Illumina Novoseq S4 lane acquiring 73 million paired-end reads (150 bp length). Two fastq files were used as input for mitogenome assembly with MITObim 1.7. The ‘-quick’ assembly option was implemented, and the mitogenome of Etheostoma nigrum (accession KT289926) was used as a reference sequence. All other parameters were left to default values. The assembly reached 500× coverage and yielded a complete genome sequence of 16,572 bp. The mitogenome was annotated with the MitoAnnotator online server (Iwasaki et al. Citation2013) and uploaded to NCBI (accession MN792799). Twenty-seven additional percidae mitogenomes were aligned using the MAFFT online server (Katoh et al. Citation2019). Alignments were checked by eye in Bioedit, revealing a gene order and GC content (45.7%) that is consistent with the mitogenomes of related species (Hall Citation1999; Huang et al. Citation2017; Jones et al. Citation2019; Kral and Watson Citation2019 Kumar et al. Citation2016). A maximum likelihood phylogenetic analysis was conducted in RAxML 8.1.12 using the GTR + G substitution model (Stamatakis Citation2014). The resulting phylogeny resolved the monophyly of Sander, Perca, Percina, and Etheostoma (). Within Etheostoma, relationships were generally poorly supported, but are comparable with previous studies (Lang and Mayden Citation2007; Near et al. Citation2011). Strong support was provided for a common ancestor between E. trisella and Etheostoma jessiae (subgenus Gemmaperca). Pairwise sequence comparisons revealed E. trisella to differ from congeners by an average of 15.2%, providing ample possibilities for developing specific eDNA protocols.
Figure 1. The evolutionary history of Percidae mitogenomes was inferred by using the Maximum Likelihood method based on the General Time Reversible model (GTR + I + G). The tree with the highest log likelihood score (−123285.9361) is shown. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.1958)). All positions containing gaps and missing data were eliminated. There were a total of 16,419 positions in the final dataset. Evolutionary analyses were conducted in MEGA7 (Kumar et al. 2016).
Acknowledgements
We thank Dominique Dawson and John Larrimore for laboratory assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Huang T, Bi G, Han Y. 2017. The complete mitochondrial genome of Etheostoma okaloosae (Perciformes: Percidae). Conservation Genet Resour. 9(4):591–593.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540.
- Jones KD, Kuhajda BR, Sandel MW. 2019. Complete mitochondrial genome for the Mobile River Basin endemic Coal Darter, Percina brevicauda (Perciformes, Percidae). Mitochondrial DNA Part B. 4(1):2031–2032.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Kral LG, Watson S. 2019. Preliminary assessment of adaptive evolution of mitochondrial protein coding genes in darters (Percidae: Etheostomatinae). F1000Res. 8:464.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Lang NJ, Mayden RL. 2007. Systematics of the subgenus Oligocephalus (Teleostei: Percidae: Etheostoma) with complete subgeneric sampling of the genus Etheostoma. Mol Phylogenet Evol. 43(2):605–615.
- NatureServe. 2013. Etheostoma trisella. The IUCN Red List of Threatened Species 2013: e.T8131A18236094. [accessed 2019 Dec 5]. 10.2305/IUCN.UK.2013-1.RLTS.T8131A18236094.en.
- Near TJ, Bossu CM, Bradburd GS, Carlson RL, Harrington RC, Hollingsworth PR, Jr Keck BP, Etnier DA. 2011. Phylogeny and temporal diversification of darters (Percidae: Etheostomatinae). Syst Biol. 60(5):565–595.
- Piggott MP. 2016. Evaluating the effects of laboratory protocols on eDNA detection probability for an endangered freshwater fish. Ecol Evol. 6(9):2739–2750.
- Ryon MG. 1986. The life history and ecology of Etheostoma trisella (Pisces: Percidae). Am Midland Naturalist. 115(1):73–86.
- Schroeter JC, Maloy AP, Rees CB, Bartron ML. 2019. Fish mitochondrial genome sequencing: expanding genetic resources to support species detection and biodiversity monitoring using environmental DNA. Conserv Genet Resour. 1–4.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
