Abstract
We performed high-throughput sequencing on the complete mitogenomes of two Araneidae species, a wasp spider (Argiope bruennichi) and a large nocturnal spider (Araneus ventricosus). The two mitogenomes contain 14,060 bp and 14,607 bp, and GC content of 26.6% and 26.5%, respectively. The two mitogenomes contain 13 protein-coding genes (PCGs), 22 transfer RNAs, 2 ribosomal RNAs, and 1 control region. We performed a phylogenetic analysis with the concatenated 13 PCGs, which showed that Argiope and Araneus are clearly classified and clustered within Araneidae clade. Our study serves to enrich evolutionary and ecological studies.
The two orb-weaver spiders, Argiope bruennichi and Araneus ventricosus, belong to the family Araneidae and are often found in forests, fields, and gardens (Krehensinkel and Tautz 2013; Kono et al. 2019). Although, the two spiders are distributed throughout Korea, data on their genetic information are lacking. In this study, we determined mitochondrial genomes of the two species. To the best of our knowledge, this is the first mitogenome report of A. ventricosus.
The two spiders, A. bruennichi (IN5987) and A. ventricosus (IN5988), were collected near Jiri mountain (35˚18′57.74″N, 127˚27′39.57″E and 35˚19′29.54″N, 127˚17′59.36″E) in Gurye-gun, Jeollanam-do, South Korea, and stored in National Institute of Biological Resources (NIBR) at Incheon, South Korea. Genomic DNA was extracted from tissue using a DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA), in accordance with the manufacturer’s protocol and deposited in the NIBR. DNA Link Inc. (Seoul, South Korea) performed whole genome resequencing with the Illumina HiSeq 2500 platform. The mitogenome assembly using the MITObim (Hahn et al. Citation2013) recovered the mitochondrial genome as a single contig with whole genome sequencing data. Genome annotations were performed using GeSeq (Tillich et al. Citation2017) with NCBI reference sequences.
Mitogenome length and AT content for A. bruennichi are 14,060 bp and 73.4%, respectively (GenBank accession: MN603962) and those for A. ventricosus, 14,607 bp and 73.5% (MN603963). The AT contents were considerably higher than the GC contents in both species. The mitogenome of both species contains 13 protein-coding genes (PCGs), 22 transfer RNA genes, 2 ribosomal RNA genes, and 1 putative control region that are typically present in metazoan mitogenomes (Boore Citation1999). The mitogenome of A. bruennichi have five start codons for 13 PCGs: ATT for ND2, ATP8, ND3, ND5, ND4L, CYTB and ND1; ATG for ATP6 and ND6; ATA for ND4; TTA for COI; and TTG for COII and COIII. Araneus ventricosus also have five initiation codons: ATT for ND2, ATP8, ND3, ND5, ND4L, CYTB and ND1; ATG for ATP6; ATA for ND4; TTA for COI; and TTG for COII, COIII and ND6. In the mitogenome of A. bruennichi, three PCGs (ND1, ND2 and CYTB) are terminated with TAG, nine PCGs (COI, COII, ATP6, ATP8, ND3, ND4, ND4L, ND5 and ND6) have TAA as a stop codon, and COIII has an incomplete codon T. Alternatively, A. ventricosus has identical codon composition except that COIII has TAA as a stop codon and ND4 ends with an incomplete T.
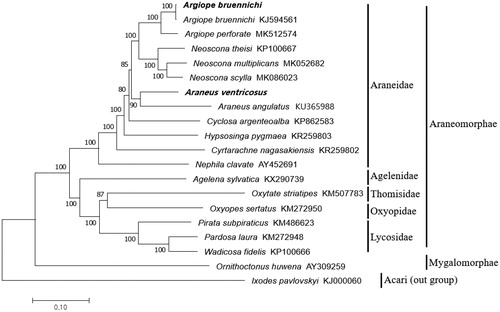
We constructed a neighbor-joining tree using MEGA7 (Kumar et al. Citation2016), and the concatenated 13 PCGs sequenced, along with 18 Arachnida mitochondrial genomes obtained from the GenBank (). Phylogenetic analysis confirmed the monophyly of Araneidae. In conclusion, this study provides two additions to the Araneae mitochondrial genome database, thus enriching in evolutionary and ecological studies.
Figure 1. Phylogenetic tree based on concatenated coding genes from complete mitochondrial genomes of 20 spiders. The sequences were obtained from Genbank and the phylogenetic tree was constructed by neighbor-joining method with 1000 bootstrap replicates. Ixodes pavlovskyi was used as an outgroup. The mitochondrial genome of A. bruennichi and A. ventricosus in bold were determined and characterized in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Boore JL. 1999. Survey and summary: animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones E, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
