Abstract
Pennisetum villosum is an ornamental grass and to better determine its phylogenetic location with respect to the other taxa in the tribe Paniceae, the complete plastid genome was sequenced. The whole plastid genome is 138,570 bp in length, consisting of a pair of inverted repeat (IR) regions of 20,269 bp, one large single-copy (LSC) region of 85,662 bp, and one small single-copy (SSC) region of 12,370 bp. The overall G + C content of the whole plastome is 38.60%. Further, maximum likelihood phylogenetic analysis with TVM + F+R2 model was conducted using 16 complete plastomes of the Poaceae, which support the close relationship between P. villosum and the species of Cenchrus L.
Pennisetum villosum as an ornamental grass is naturally distributed on a hilly area in warmer regions of Africa in the family Poaceae (http://foc.iplant.cn/). Several species of Pennisetum are popular in the garden for their bottlebrush spikes and cascading foliage, such as P. alopecuroides (L.) Spreng., P. setaceum (Forssk.) Chiov., and P. villosum Fresen. (Crins Citation1991; Rúgolo de Agrasar and Puglia Citation2004). For a better understanding of the relationships of P. villosum and other taxa in the tribe Paniceae, it is necessary to reconstruct a phylogenetic tree based on high-throughput sequencing approaches.
About 5 cm long leaves of P. villosum in Kunming Yiliang county (Yunnan, China; 103˚8″51.21″ E, Lat. 24˚55′31.03″ N, alt. 1636 m) were collected for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group of Xishuangbanna Tropical Botanical Garden (HITBC: accession number: HFTC-Qi Wang 001). The complete chloroplast genome was sequenced following Zhang et al. (Citation2016) and their 15 universal primer pairs were used to perform long-range PCR for next-generation sequencing. Clean paired-end reads were assembled with the Get Organelle Kit (version 1.4.0) (Jin et al. Citation2018). The contigs were aligned using the publicly available plastid genome of Cenchrus purpureus (GenBank accession number MF594682) (Wu et al. Citation2019) and annotated in Geneious 4.8.
The plastome of P. villosum (SY001902) with a length of 138,570, was 398 bp and 371 bp larger than that of Cenchrus americanus (138,172 bp, KX756179) and C. purpureus (138,199 bp, MF594682) (Wu et al. Citation2019). The length of the inverted repeats (IRs), large single-copy (LSC), and small single-copy (SSC) regions of P. villosum was 20,269 bp, 85,662 bp, and 12,370 bp, respectively. The overall G + C content was 38.60%.
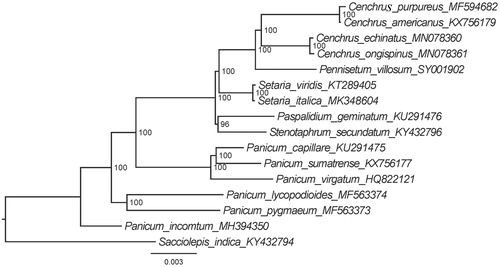
Based on 16 published complete chloroplast genome sequences, we reconstructed a phylogenetic tree () to confirm the relationship between P. villosum and other related taxa with published plastomes in Cenchrus L., Panicum L., Paspalidium Stapf, Setaria P. Beauv., and Stenotaphrum Trin., while Sacciolepis indica (L.) Chase as outgroup. Maximum likelihood (ML) phylogenetic analyses were performed base on TVM + F+R2 model in the iqtree version 1.6.7 program with 1000 bootstrap replicates (Nguyen et al. Citation2015). The ML phylogenetic tree with 99–100% bootstrap values at each node supported that P. villosum and four taxa of Cenchrus were located in the same clade, suggesting sisterhood of the P. villosum and the species of Cenchrus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data archiving statement
The plastome data of the Pennisetum villosum will be submitted to Chloroplast Genome Database (https://lcgdb.wordpress.com). Accession numbers are SY001902.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Crins WJ. 1991. The genera of Paniceae (Gramineae: Panicoideae) in the Southeastern United States. J Arnold Arbor. 1(Suppl. 1):205–220.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722.
- Jin JJ, Yu WB, Yang JB, Song Y, Yi TS, Li DZ. 2018. GetOrganelle: a simple and fast pipeline for de novo assembly of a complete circular chloroplast genome using genome skimming data. bioRxiv. 156479.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Rúgolo de Agrasar ZE, Puglia ML. 2004. Gramíneas ornamentales. In: Hurrel JA, editor. Plantas de la Argentina: silvestres y cultivadas. Buenos Aires: Editorial LOLA; pp. 1–336.
- Wu D, Wang Y, Zhang L. 2019. The complete chloroplast genome sequence of Cenchrus purpureus. Mitochondr DNA B. 4(2):3560–1734.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J System Evol. 54(3):219–227.