Abstract
We determined the complete mitochondrial genome (mitogenome) of the leafhopper Idiocerus herrichii by next-generation sequencing. Similar to the typical mtDNA of insects, the mitogenome is 15,489 bp in size, with an A + T content of 78.4%, comprises 13 protein-coding genes (PCGs), two rRNA genes, 22 tRNA genes, and one non-coding region. Most of the PCGs are initiated with the typical ATN codon, with exception to ATP8 and ND5, which are started with TTG. Most PCGs are terminated with the typical TAA codon, except for COX2, ND5, ND4 and ND1 with the incomplete termination codon T. The concatenated PCGs were used to conduct Bayesian phylogenetic analyses together with 20 mitogenome data of leafhoppers in GenBank. The phylogenetic tree confirms the monophyly of Idiocerinae, indicating I. herrichii being closely related to I. laurifoliae plus Populicerus popili.
The idiocerine leafhoppers, i.e. Idiocerinae, represent one of the largest groups of leafhoppers with more than 800 species in 106 genera, occurring in all zoogeographical regions (Zhang and Webb Citation2019). In this subfamily, there are approximately 91 species belonging to 26 genera known from China (Xue et al. Citation2016; Zhang and Wang Citation2018). To date, only five complete mitochondrial genomes have been sequenced for the Idiocerinae subfamily (Wang et al. Citation2018).
In this study, we sequenced and annotated the complete mitochondrial genome (mitogeonome) of Idiocerus herrichii, a Western Palearctic species of the idiocerine leafhoppers. The specimens were collected from the Awei beach, Xinjiang Uygur Autonomous Region, China on 9 May 2019, which were selected as specimen (number IMNU201905040001). The entire body without abdomen was shipped to Tsingke (Beijing, China) for genomic extraction and 150-base-pair paired-end library construction; sequencing was performed on an Illumina HiSeq 2000 instrument. De novo assembly of clean reads was performed using SPAdes v3.11.0 (Bankevich et al. Citation2012). The mitogenome of Idiocerus laurifolidae (GenBank accession number NC_089741) was further used as a reference to assemble the sequenced sample. Genes were annotated with the MITOS (Bernt et al. Citation2013) web server.
The complete mitogenome of I. herrichii (GenBank accession number MN935487) is 15,489 bp in length, containing 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one large non-coding [A + T]-rich region. The gene content, arrangement, and composition exhibited a typical insect mitogenome feature. Overall, the I. herrichii mitogenome has an A + T content of 78.4% (A 43.2%; T 35.2%; C 11.8%; G 9.8%), which is within the range reported for hemipteran mitogenomes (Zhang et al. Citation2014). The majority of the genes in the mtDNA of I. herrichii is distributed on H-strand, except for the four PCGs (ND5, ND4, ND4L and ND1), two rRNA (16S and 12S) and eight tRNA genes (tRNA-Gln, Cys, Tyr, Phe, His, Pro, Leu2 and Val). Most of the PCGs are initiated with the typical ATN codon, except for ATP8 and ND5 with TTG. Most PCGs are terminated with the typical TAA codon, except for COX2, ND5, ND4 and ND1 with the incomplete termination codon T. The 16S rRNA gene is 1203 bp in size and is located between tRNA-Leu2 and tRNA-Val; the 12S rRNA gene is 754 bp in length and is located after tRNA-Val. The non-coding region is 1171 bp long, and is located between 12S rRNA and tRNA-Ile.
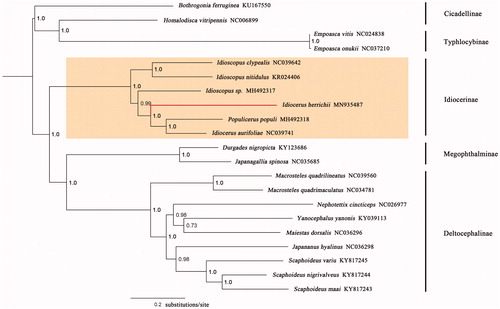
The phylogenetic analyses of I. herrichii and other 20 leafhopper species were conducted using the concatenated nucleotide sequences of the 13 PCGs. Sequences were aligned with MEGA6 software (Tamura et al. Citation2013). Phylogenetic trees were generated using MrBayes v3.2.1 (Ronquist et al. Citation2012) with GTR + G substitution model. The resulting phylogenetic tree confirmed the monophyly of Idiocerinae, Deltocephalinae, Typhlocybinae, and Megophthalminae with robust posterior probabilities, respectively (). Idiocerus herrichii is shown to be closely related to I. laurifoliae plus Populicerus popili. The mitogenome obtained herein will contribute to the examination of phylogenetic relationships and systematics for the idiocerine leafhoppers.
Figure 1. A majority-rule consensus tree (midpoint rooted) inferred from Bayesian inference using MrBayes v3.2.1 under the GTR + G model, based on the concatenated PCGs. Node numbers show Bayesian posterior probabilities. Branch lengths represent means of the posterior distribution. GenBank accession numbers are given with species names.
Acknowledgements
We would like to express our sincere gratitude to Dr. Xianguang Guo (Chengdu Institute of Biology, Chinese Academy of Sciences) for his help with revising the manuscript. We also thank Minli Chen for her helpful suggestions.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wang JJ, Yang MF, Dai RH, Li H, Wang XY. 2018. Characterization and phylogenetic implications of the complete mitochondrial genome of Idiocerinae (Hemiptera: Cicadellidae). Int J Biol Macromol. 120:2366–2372.
- Xue QQ, Viraktamath CA, Zhang YL. 2016. Checklist to Chinese Idiocerinae leafhoppers, key to genera and description of a new species of Anidiocerus (Hemiptera: Auchenorrhyncha: Cicadellidae). Entomol Am. 122:405–417.
- Zhang B, Wang CF. 2018. Henanocerus lineatus gen. nov. and sp. nov., a new leafhopper feeding on Populus tomentosa Carrière from China (Hemiptera, Cicadellidae). Zootaxa. 4442(4):584–588.
- Zhang B, Webb MD. 2019. A checklist and key to the idiocerine leafhoppers (Hemiptera: Cicadellidae) of Hainan Island, with description of a new genus and new species. Zootaxa. 4576(3):581–587.
- Zhang KJ, Zhu WC, Rong X, Liu J, Ding XL, Hong XY. 2014. The complete mitochondrial genome sequence of Sogatella furcifera (Horvath) and a comparative mitogenomic analysis of three predominant rice planthoppers. Gene. 533(1):100–109.
