Abstract
The mitochondrial genome of Paradoxornis heudei is described in this study. The molecule is 16,924 bp in length and contains 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and an AT-rich region. All PCGs use the typical start and stop codons, except COX3, ND2, and ND4 use T or TA as their stop codons. The rrnL and rrnS genes are 1602 bp and 984 bp in length, respectively. Phylogenetic analysis shows that the least evolved Paradoxornis locates at the basic position of Muscicapidae and among Paradoxornis, the other three species are firstly clustered together, then constitute a monophyletic group with P. heudei.
The reed parrotbill Paradoxornis heudei belongs to the genus Paradoxornis of Muscicapidae, is endemic to eastern China and southeastern Siberia (BirdLife International Citation2017). Because of the loss and degradation of reedbed habitats, the species was rapidly declined in recent years and was listed as NT (Near Threatened) in IUCN red list (BirdLife International Citation2017). Up to date, the taxonomic status of P. heudei is still standing as a controversial issue and need to be elucidated. In this study, we sequenced and analyzed the complete mitochondrial genome of P. heudei to provide insights into the phylogenetic relationship of Muscicapidae.
Adult individuals of P. heudei were collected at the Dafeng Milu National Reserve, Jiangsu province, China (coordinates: N33°05′, E120°49′). After morphological identification, fresh samples were preserved in 100% ethanol and kept in the zoological museum (NJFU-201907, Nanjing Forestry University, China) at 20 °C until use. Total genomic DNA was extracted from the thorax muscle of a single individual using the Sangon Animal genome DNA Extraction Kit (Shanghai, China). The constructed library was quantified by Qubit v3.0 (Life Technologies, Carlsbad, US) and then sequenced on the Illumina HiSeq X sequencing platform. The resultant reads were assembled and annotated using the SPAdes v3.11.1 (Bankevich et al. Citation2012) and MITOS software (Bernt et al. Citation2013).
The whole mitogenome is a circular molecule of 16,924 bp in size (GenBank accession No. MN865117), and contains 13 protein-coding genes (PCGs), 22 transfer tRNA genes, and 2 ribosomal rRNA genes, along with an AT-rich control region. Its gene arrangement and orientation are identical to those of other known Paradoxornis mitogenomes (Zhang et al. Citation2015; Wen et al. Citation2017; He et al. Citation2019; Zhou et al. Citation2019). The A + T content of the mitogenome is 54.1%, which is generally in accordance with other Paradoxornis mitogenomes. Resembling typical avian mtDNA, all mitochondrial genes of P. heudei are encoded on the H-strand, apart from one PCG (ND6) and eight tRNAs (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UGA), tRNAPro, and tRNAGlu) (Dove et al. Citation2008). All PCGs are initiated by the typical ATG codon. Ten PCGs have typical stop codons (TAA/TAG/AGA/AGG), whereas three PCGs (COX3, ND2 and ND4) stop with incomplete TA or T. The rrnL and rrnS genes are 1602 bp and 984 bp in length, respectively.
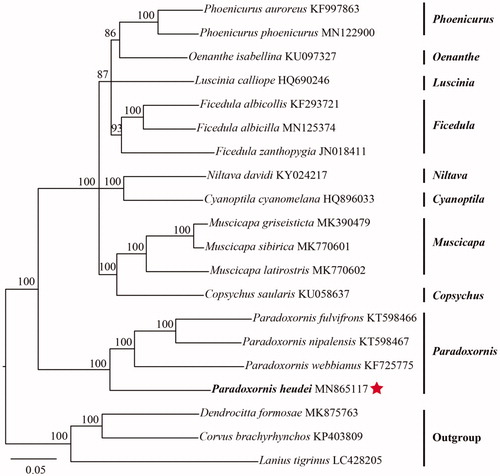
Using Corvus brachyrhynchos, Dendrocitta formosae, and Lanius tigrinus as outgroups, we performed the maximum-likelihood (ML) phylogenetic analysis to clarify the phylogeny of 17 Muscicapidae species, including the P. heudei, based on concatenated nucleotide sequences of 13 PCGs with PhyML v3.7 (Guindon et al. Citation2010). The resultant ML tree shows that all chosen Muscicapidae species include 9 genera and the least evolved Paradoxornis locates at the basic position of Muscicapidae (). Among Paradoxornis, P. fulvifrons, P. nipalensis, and P. webbianus are firstly clustered together, then constitute a monophyletic group with P. heudei ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF, 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- BirdLife International. 2017. Paradoxornis heudei. The IUCN Red List of Threatened Species 2017: e.T22716844A111640218. 10.2305/IUCN.UK.2017-1.RLTS.T22716844A111640218.en
- Dove CJ, Rotzel NC, Heacker M, Weigt LA, 2008. Using DNA barcodes to identify bird species involved in birdstrikes. J Wildlife Manage. 72(5):1231–1236.
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O, 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. System Biol. 59(3):307–321.
- He W, Xu H, Li D, Xie M, Zhang M, Ni Q, Yao Y, 2019. Complete mitochondrial genome sequence of grey-headed parrotbill (Paradoxornis gularis) and its phylogenetic analysis. Mitochondr DNA B-Resour. 4(2):3669–3670.
- Wen L, Yang X, Liao J, Fu Y, Dai B, 2017. The complete mitochondrial genome of the fulvous parrotbill Paradoxornis fulvifrons (Passeriformes: Muscicapidae). Mitochondr DNA A. 28(1):143–144.
- Zhang H, Li Y, Wu X, Xue H, Yan P, Wu X-B, 2015. The complete mitochondrial genome of Paradoxornis webbianus (Passeriformes, Muscicapidae). Mitochondrial DNA. 26(6):879–880.
- Zhou C, Jin J, Chen Y, Hao Y, Zhang X, Meng Y, Yang N, Yue B, 2019. Two new complete mitochondrial genomes (Paradoxornis gularis and Niltava davidi) and their phylogenetic and taxonomic implications. Mitochondr DNA B Resour. 4(1):820–821.