Abstract
The mitochondrial genome of one potamidae species Neilupotamon papileonaceum was sequenced and annotated. The mitogenome is 16,273 bp in length, containing 37 typical genes. The A + T content of the whole mitogenome is 68.9%. Most of the PCGs started with ATG and stopped with TAT, except for ATP8 and ND6 stopped with TGA, COX1, COX2, ND5, ND4, and ND4L used incomplete T as stop codon. The phylogeny tree is monophyletic among 15 related species. The relationships of N. papileonaceum, Sinopotamon yaanense, and Longpotamon yangtsekiense were closer than others. This study further enriched mitogenome database of the tribe Typhlocybini.
The Neilupotamon papileonaceum as a kind of freshwater crab belongs to Crustacea, Decapoda, Potamidae. The N. papileonaceum is distributed in Huitong County, Loudi County, Xinhua County, and Chenxi County, Hunan province, China. Their whole body is red with blue butterfly patterns on the carapace and live in caves in streams and canals (Dai Citation1999).
A male adult of N. papileonaceum was collected from Shuiche Town(Xinhua County, Hunan Province, China) (110°97.5′E, 27°69.9′N), in August 2018 and the specimens (No. DWNLXX-1) were stored at −80 °C in our laboratory (College of Animal Science and Technology, Hunan Agriculture University). Total genomic DNA of the N. papileonaceum was extracted from the muscle of cheliped using the Column Animal DNA out (Beijing Tiandz, Beijin, China) following the kit’s instructions. Then, the mitochondrial genome (mitogenome) of N. papileonaceum was sequenced by Illumina sequencing platform (Wuhan Biowefind Biotechnology Co., Ltd, Wuhan, China). The reads were assembled and annotated using Generous Prime (v2019.1.3.). All tRNA genes were identified by ARWEN v1.2 (Laslett and Canbäck Citation2008). The annotated sequences of mitogenome were submitted to GenBank with accession number MT021974. All protein condoning genes (PCGs) were aligned using MAFFT algorithm in the TranslatorX (Katoh et al. Citation2019), and then poorly aligned results were removed by Gblocks 9.1 b (Castresana Citation2000; Abascal et al. Citation2010).
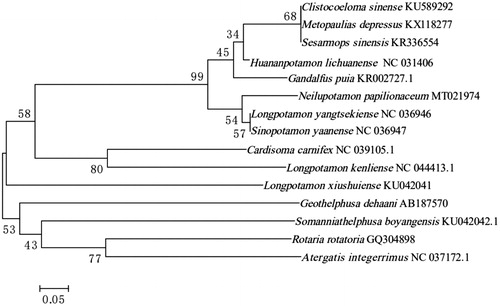
Mitogenome of N. papileonaceum has 16,273 bp in length and including 37 typical genes (13 PCGs, 22 tRNA genes, and 2 rRNA genes). The overall nucleotide composition of the mitogenome is 34.1% for A, 21.5% for C, 9.7% for G, and 34.7% for T. The A + T content of genes is 68.9%. Most PCGs started with ATG and stopped with TAT, except for ATP8 and ND6 stopped with TGA, COX1, COX2, ND5, ND4, and ND4L used incomplete T as stop codon. The length of 22 tRNA ranges from 62 bp (tRNA-Gly and tRNA-Cys) to 72 bp (tRNA-Val). Genes of 16S rRNA and 12S rRNA are 1331 bp and 883 bp, respectively. To validate the phylogenetic position of N. papileonaceum, we perform multiple sequence alignment and MEGA 6.0 (Tamura et al. Citation2013) to construct a neighbor-joining tree containing complete mitochondrial genome DNA of 15 species in Decapoda, as shown in the phylogenetic tree ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequence guided by amino acid translations. Nucleic Acids Res. 38(suppl_2):W7–W13.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol E. 17(4):540–552.
- Dai A. 1999. Animal of China, Arthropoda, Subphylum crustaceans, Malacostraca, Decapodas, Parathelphusicae, Potamoidea. Beijing: Science Press; p. 374–376.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequence. Bioinform. 24(2):172–175.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol E. 30(12):2725–2729.