Abstract
Chromolaena odorata is a perennial noxious invasive weed to south China. In this study, we assembled the complete chloroplast (cp) genome of C. odorata by next-generation sequencing technologies. The whole cp genome is 151,270 bp in size, consisting of a pair of inverted repeats (IR 25,080 bp), a large single-copy region (LSC 82,663 bp), and a small single-copy region (SSC 18,447 bp). The genome contains a total of 132 genes, including 88 protein-coding genes, 36 tRNAs, and 8 rRNAs. Furthermore, a maximum likelihood phylogenetic analysis demonstrated that C. odorata was closely related to Praxelis clematidea and Ageratina adenophora. The cp genome will provide reference for the further investigation and research of C. odorata.
Chromolaena odorata is one of the most serious 100 invasive alien species in the world (Lowe et al. Citation2000). It is an extremely fast-growing, high-reproducing, and long-lived perennial herbaceous shrub of Compositae family native to Central and South America (Yu et al. Citation2016). In China, C. odorata was first discovered in Yunnan and Hainan provinces in 1934, and now it is rapidly spreading throughout the southern China. It not only affects the production of agriculture, forestry and husbandry and causes great economic loss, but also threatens the health of human and livestock (Yu et al. Citation2010). Currently, studies on C. odorata focus in its invasion mechanism, ecological impacts and control methods, such as phenotypic plasticity (Liao et al. Citation2019), genetic variation (Paterson and Zachariades Citation2013), plant–soil interaction (Te Beest et al. Citation2015), biodiversity (Rozen-Rechels et al. Citation2017), and biological control (Aigbedion-Atalor et al. Citation2019). However, its phylogenetic relationships are rather limited. In this study, we reported the complete chloroplast (cp) genome sequence of C. odorata, which would be helpful for its genetic and evolutionary research.
The leaves of C. odorata were collected from the suburb of Yangjiang city (21°89’76.35″N, 112°00’56.63″E), Guangdong province, China. The specimen was stored at Guangzhou City Polytechnic (specimen code GCP0636). The genomic DNA of C. odorata was extracted from leaves by plant genomic DNA kit (Tiangen Biotech, China) and sequenced using the Novaseq platform (Illumina, San Diego, CA) following the manufacturer’s recommendations. The plastome was assembled by the GetOrganelle (Jin et al. Citation2019) and annotated with the Geseq (Tillich et al. Citation2017). Finally, a complete chloroplast genome of C. odorata was obtained and submitted to GenBank with the accession number MN885889.
The complete cp genome of C. odorata is 151,270 bp in length, containing a large single-copy (LSC) region of 82,663 bp, a small single-copy (SSC) region of 18,447 bp, and two inverted repeat (IR) regions of 25,080 bp. The new sequence has 132 genes in total, including 88 protein-coding genes, 36 tRNA genes, and 8 rRNA genes. In addition, the overall GC content of the genome is 37.48%, whereas the corresponding values of the LSC, SSC, and IR regions are 54.65%, 12.19%, and 16.58%, respectively.
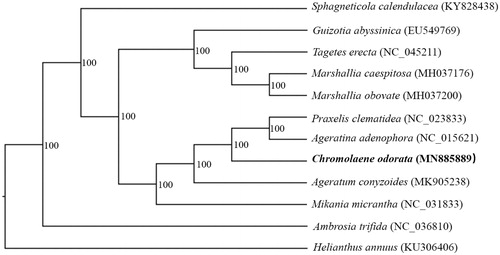
To further investigate its phylogenetic position, 12 complete cp genomes of Compositae species (Sphagneticola calendulacea, Guizotia abyssinica, Tagetes erecta, Marshallia caespitosa, Marshallia obovate, Praxelis clematidea, Ageratina adenophora, Chromolaena odorata, Ageratum conyhzoides, Mikania micrantha, Ambrosia trifida, Helianthus annuus) were aligned using MAFFT (Katoh and Standley Citation2013). All the data were downloaded from NCBI GenBank. A maximum likelihood analysis was performed by RAxML (Stamatakis Citation2014) with 1000 bootstrap replicates (Minh et al. Citation2013; Chernomor et al. Citation2016). The results showed that C. odorata was closely related to Praxelis clematidea and Ageratina adenophora with 100% bootstrap support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Aigbedion-Atalor PO, Day MD, Idemudia I, Wilson DD, Paterson ID. 2019. With or without you: stem-galling of a tephritid fly reduces the vegetative and reproductive performance of the invasive plant Chromolaena odorata (Asteraceae) both alone and in combination with another agent. BioControl. 64(1):103–114.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2019. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. bioRxiv. doi: 10.1101/256479.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol E. 30(4):772–780.
- Liao Z-Y, Scheepens JF, Li W-T, Wang R-F, Zheng Y-L, Feng Y-L. 2019. Biomass reallocation and increased plasticity might contribute to successful invasion of Chromolaena odorata. Flora. 256:79–84.
- Lowe S, Browne M, Boudjelas S, De Poorter M. 2000. 100 of the world’s worst invasive alien species: a selection from the global invasive species databases. Auckland, New Zealand: The Invasive Species Specialist Group (ISSG) of the Species Survival Commission (SSC) of the World Conservation Union (IUCN).
- Minh BQ, Nguyen MAT, von Haeseler A. 2013. Ultrafast approximation for phylogenetic bootstrap. Mol Biol E. 30(5):1188–1195.
- Paterson ID, Zachariades C. 2013. ISSRs indicate that Chromolaena odorata invading southern Africa originates in Jamaica or Cuba. Biol Control. 66(2):132–139.
- Rozen-Rechels D, Te Beest M, Dew LA, Le Roux E, Druce DJ, Cromsigt J. 2017. Contrasting impacts of an alien invasive shrub on mammalian savanna herbivores revealed on a landscape scale. Diversity Distrib. 23(6):656–666.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Te Beest M, Esler KJ, Richardson DM. 2015. Linking functional traits to impacts of invasive plant species: a case study. Plant Ecol. 216(2):293–305.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11.
- Yu FK, Akin-Fajiye M, Thapa Magar K, Ren J, Gurevitch J. 2016. A global systematic review of ecological field studies on two major invasive plant species, Ageratina adenophora and Chromolaena odorata. Diversity Distrib. 22(11):1174–1185.
- Yu XQ, Feng YL, Li QM. 2010. Review of research advances and prospects of invasive Chromolaena odorata. Chin J Plant Ecol. 34(5):591–600.