Abstract
The complete mitochondrial genome of Holothuria edulis was obtained and described in this study. This complete mitochondrial genome is 15,743 bp in length and consists of 13 protein-coding genes, 2 ribosomal RNA genes, and 22 transfer RNA genes. Except ND6 and 5 tRNAs, the others were encoded by the heavy strand. The overall base composition of the heavy-strand was 32.29% A, 15.25% G, 25.88% C, and 26.57% T, with a high A + T content of 58.86%. The phylogenetic analysis suggested that H. edulis was closest to H. pervicax. The newly described mitochondrial genome may provide valuable data for phylogenetic analysis for Holothuroidea.
Holothuroidea, as known as sea cucumber, has important economical value due to high nutrition (Wen et al. Citation2010). Holothuria edulis is a widely distributed sea cucumber species in most parts of the Indo-Pacific region (Liao Citation1997). It was found in a variety of habitats, including rocky reefs, coral reefs, seagrass, and mudflats (Conand Citation2008). Both sexual reproduction and asexual reproduction of H. edulis were observed in nature (Uthicke Citation1997). So far, there are only a few DNA sequences such as COX1 and 16S rRNA of H. edulis were reported (Soliman et al. Citation2016). Lack of genetic resources has hindered conservation and utilization of H. edulis. Mitochondrial DNA is an effective tool for species identification, molecular taxonomy, and population genetic analyses (Galtier et al. Citation2009). In this study, the complete mitochondrial genome of H.edulis and its phylogenetic relationships within other sea cucumbers were investigated.
The specimen was collected from Shenzhen, Guangdong province, China (N22°35′, E114°31′) and kept in the Department of Biology, Lingnan Normal University (Zhanjiang, Guangdong province, China). The dorsal body muscle of H.edulis were fixed in 100% ethanol and stored at −20 °C. After sampling, the whole body specimen were stored in 100% ethanol and deposited at the Zoological Herbarium, Lingnan Normal University (Acc. Number SC20190818-5).
Approximately 30 mg of muscle tissue was used for mitochondrial DNA (mtDNA) extraction with TIANamp Marine Animals DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s specification. MtDNA were sequenced using the Illumina Hiseq Sequencing System (Illumina Inc., San Diego, CA). The clean data were acquired and assembled by the SPAdes and PRICE (Bankevich et al. Citation2012). BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) and ORFs finder https://www.ncbi.nlm.nih.gov/orffinder/) were used to identify and annotate protein-coding genes (PCGs). tRNAscan-SE 1.31 (Lowe and Eddy Citation1997) was used to identify tRNA genes. A phylogenetic tree was constructed using MEGA 6.0 software.
The mitochondrial genome of H. edulis is 15,743 bp in length (GenBank accession number: MT084774), containing the typical set of 13 PCGs, 22 tRNA and 2 rRNA genes, and a putative control region. The heavy strand consists of 32.29% A, 25.88% C, 15.25% G and 26.57% T bases, with a high A + T content of 58.86%. Of the 37 genes, 31 were encoded by the heavy strand, and the others were encoded by the light strand. All PCGs were initiated by ATG. Eleven PCGs were terminated with the typical TAA or TAG as stop codon, while cox2 and ND4 ended with T––. Twenty-two tRNA genes ranged in size from 61 to 73 bp.
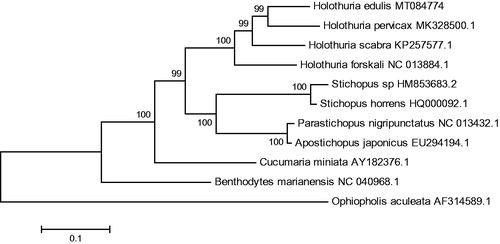
The phylogenetic tree was constructed based on 13 concatenated PCGs from 10 sea cucumbers from Genbank database by the maximum likelihood (ML) method. Ophiopholis aculeata was used as an outgroup for tree rooting (). It was demonstrated that H. edulis was clustered with H. pervicax, which may suggest that the H. edulis were most closely related to the H. pervicax. In all, this genome will contribute to future phylogenetic studies and population genetic analyses for H.edulis.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19:455–477.
- Conand C. 2008. Population status, fisheries and trade of sea cucumbers in Africa and Indian Ocean. In Toral-Granda V, Lovatelli A, Vasconcellos M, editors. Sea cucumbers. A global review on fishery and trade. FAO Fisheries Technical Paper No. 516. Rome: FAO; p. 153–205.
- Galtier N, Nabholz B, Glemin S, Hurst G. 2009. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol Ecol. 18(22):4541–4550.
- Liao Y. 1997. Fauna sincia: phylum Echinodermata; class Holothuroidea. Beijing: Science Press.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucleic Acids Res. 25(5):955–964.
- Soliman T, Fernandez-Silva I, Reimer JD. 2016. Genetic population structure and low genetic diversity in the over-exploited sea cucumber Holothuria edulis Lesson, 1830 (Echinodermata: Holothuroidea) in Okinawa Island. Conserv Genet. 17(4):811–821.
- Uthicke S. 1997. The seasonality of asexual reproduction in Holothuria (Halodeima) atra, Holothuria (Halodeima) edulis and Stichopus chloronotus (Holothuroidea: Aspidochirotida) on the Great Barrier Reef. Mar Biol. 129(3):435–441.
- Wen J, Hu CQ, Fan SG. 2010. Chemical composition and nutritional quality of sea cucumbers. J Sci Food Agric. 90(14):2469–2474.