Abstract
In this study, we obtained the complete mitochondrial genome of Pseudoxenodon macrops. The sequenced mitogenome is 20,978 base pairs(bp) in length, which contained 13 protein-coding genes (PCGS), 22 transfer RNA genes (tRNA), 2 Ribosomal RNA genes (rRNA) and 2 control regions (D-loop). The total base composition of mitochondrial DNA is that A, C, G and T occupied 33.1%, 26.6%, 12.8% and 26.5%, respectively. The mitochondrial genome of Pseudoxenodon macrops will help us reveal phylogenetic relationships among the species of the Colubridae family.
Pseudoxenodon macrops belongs to Colubridae family(Wang and He Citation1994), and lives in the mountainous area with an altitude of 900–1800 m(Zhao Citation2003; Bhosale and Thite Citation2013). There are three subspecies of Pseudoxenodon macrops in China, and these species are distributed in Henan, Guizhou, Hubei, Sichuan, Gansu, Fujian and Guangxi provinces(Farkas and Fritz Citation1999; Ji and Wen Citation2002). In this study, we sequenced the complete mitochondrial genome sequences of Pseudoxenodon macrops and combined with the existing mitochondrial genome sequence of Colubridae family in GenBank to construct a phylogenetic tree. These results can reveal the phylogenetic relationship between Pseudoxenodon macrops and other species in the Colubridae family.
The specimen of Pseudoxenodon macrops was collected from Mount Emei (Latitude: 29°33′11.64″N, Longitude: 103°23′16.72″E, Altitude: 910 m), and stored in the Zoological Museum(Specimen number: EM1906003), College of Life Sciences, Sichuan Normal University, China. In this study, the complete mitochondrial genome sequence was obtained by high-throughput sequencing method with Illumina Hiseq 2500 (Tsingke, Tianjin). And the complete sequence of the mitochondrial genome was submitted to GenBank.
The mitochondrial genome of Pseudoxenodon macrops is 20,978 bp in length(GenBank accession number: MN427889), which contains 13 protein-coding genes(ND1, ND2, ND3, ND4, ND4L, ND5, ND6, ATP6, ATP8, COI, COII, COIII, Cytb), 22 transfer RNA genes(tRNA), 2 ribosomal RNA genes(rRNA), and 2 control regions(D-loop). The base composition is 34.3% for A, 26.5% for C, 12.8% for G, and 26.4% for T. The large ribosomal RNA(lrRNA) is 1471 bp in length and small ribosomal RNA (srRNA) is 1138 bp in length. The lengths of the two control regions are 2325 bp and 2067 bp, respectively. ND6 and nine tRNAs are encoded by the L-strand, whereas all the other genes are encoded by the H-strand.
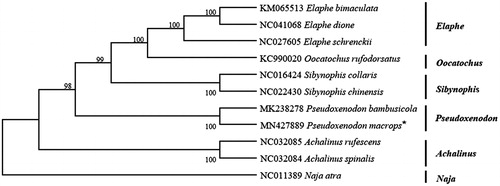
Based on the concatenated nucleotide sequences of protein-coding genes and two rRNAs, the phylogenetic relationships of the Pseudoxenodon macrops and the other 10 snakes were constructed by MEGA6.0 using maximum-likelihood(ML) method with 1000 bootstrap replications(Tamura et al. Citation2013). The phylogenetic tree () showed that the Pseudoxenodon macrops was closer to Pseudoxenodon bambusicola (genus Pseudoxenodon) in genetic relationship, which is consistent with the results of traditional morphological classification(Zhang and Huang Citation2013). This study provides data for the systematic classification of Colubridae. However, the molecular evidence inferred in this study is limited, more mitochondrial genomic information of other snakes is necessary in order to elucidate the evolutionary relationships within major lineages of Colubridae.
Figure 1. Phylogenetic tree inferred from maximum likelihood analysis of the nucleotide of protein-coding genes and two ribosomal RNA genes. Naja atra was used as outgroup. The nodal numbers indicate the bootstrap values obtained with 1000 replicates. The genbank accession number, species name and generic name are shown on the right side of the phylogenetic tree. The newly sequenced mitogenome is indicated by the asterisk.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bhosale HS, Thite V. 2013. Death feigning behavior in large-eyed false cobra Pseudoxenodon macrops (Blyth, 1854) (Squamata: Colubridae). Russ J Herpetol. 20(3):190–192.
- Farkas B, Fritz U. 1999. Geographic distribution. Pseudoxenodon macrops. Herpetol Rev. 30(3):175.
- Ji DM, Wen SS. 2002. Atlas of reptilias of China. Zhengzhou, China: Henan Science and Technology Press. (in Chinese)
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30(12):2725–2729.
- Wang RF, He WS. 1994. Study on the karyotype, high resolution banding meiosis of Pseudoxenodon macrops (snake). Zool Res. 15(3):29–32.
- Zhang BL, Huang S. 2013. Relationship of old world Pseudoxenodon and new world dipsadinae, with comments on underestimation of species diversity of Chinese Pseudoxenodon. Asian Herpetol Res. 4(3):155–165.
- Zhao EM. 2003. Coloured atlas of Sichuan reptiles. Beijing, China: China Forestry Publishing House. (in Chinese)
