Abstract
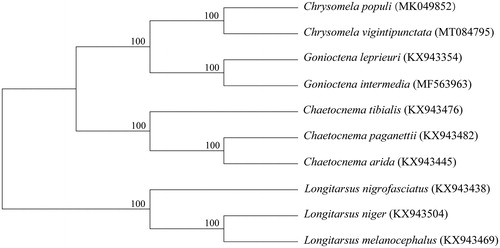
The complete mitochondrial genome of Chrysomela vigintipunctata was sequenced and reported here. The circle genome of the leaf beetle is 17,474 bp in length. There are 38 sequence elements including 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and a control region. Phylogenetic relationships of 10 Chrysomelidae species indicated by the phylogenetic tree were in agreement with their taxonomic relationships based on morphological characteristics, while the C. vigintipunctata were clustered together with C. populi.
Chrysomela vigintipunctata (Scopoli 1763) belongs to the family Chrysomelinae, and includes two subspecies, C. vigintipunctata vigintipunctata (Scopoli) and C. vigintipunctata alticola (Wang) which are apparently different in morphology. C. vigintipunctata is widely distributed across the temperate forest zone of the Palearctic (Bienkowski Citation2004)while the distributing altitude of the two subspecies is different, with the former below 3200 m and the latter mainly distributed over 3200 m (Ge et al. Citation2004). This leaf beetle is harmful to willows (Salix spp.), and can even result in severe defoliation to its host plants (Urban Citation1997).
In this study, C. vigintipunctata adults were collected from Yushu, Qinghai Province, China (33°09′26″N, 97°21′45″E) in August 2019 and were identified as the subspieces C. vigintipunctata alticola (Wang) based on the morphological feature. The specimen were kept in the Insect Collection of the Entomology Lab, College of Agriculture and Animal Husbandry, Qinghai University, Xining, China (accession number: YJY-2019-LYJ001). Genomic DNA was extracted using the commercial kit (Ezup Column Animal Genomic DNA Purification Kit, Sangon Biotech). Genomic sequencing was performed on the Illumina HiSeq Platform (Illumina, San Diego, CA) with a read length of 150 bp. The software SPAdes v.3.14.0 (Bankevich et al. Citation2012) was employed to assemble the mitogenome. The assembled sequence was then annotated using the web server MITOS (Bernt et al. Citation2013).
A circularized DNA assembly 17,474 bp in length was harvested and deposited in the GenBank with accession number of MT084795. Totally, 37 genes are recognized in this mitogenome: 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes and 2 ribosomal RNA (rRNA) genes, as well as a non-coding AT rich control region(D-loop). Of all 38 sequence elements, 4 PCGs, 9 tRNA genes and 2 rRNA genes are located on the light strand, while others are on the heavy strand.
Based on mitogenomes assembled here or downloaded from GenBank, phylogenetic relationships of 10 Chrysomelidae species were resolved by means of Neighbor-joining (). After aligned using MAFFT (Katoh and Standley Citation2013), the Neighbor-joining tree was built using MEGA7 (Kumar et al. Citation2016) with bootstrap set to 1000. Phylogenetic relationships indicated by the phylogenetic tree were in agreement with their taxonomic relationships based on morphological characteristics, while the C. vigintipunctata were clustered together with another species from the same genus, i.e., C. populi.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Bienkowski AO. 2004. Leaf-beetles (Coleoptera, Chrysomelidae) of the Eastern Europe. New key to subfamilies, genera, and species. Moscow: Mikron-print. p. 278.
- Ge S, Cui J, Yang X. 2004. Comparative morphology of two subspecies of Chrysomela vigintipunctata (Scopoli) (Coleoptera, Chrysomelidae, Chrysomelinae). Acta zootaxonomica Sinica. 29(3):415–422.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Urban J. 1997. On the occurrence, bionomics and harmfulness of Chrysomela vigintipunctata in the Czech Republic. Lesnictvi-UZPI (Czech Republic).