Abstract
Usnea is a genus of lichenized Ascomycete that grows in moist mountain areas. It is a food for wild animals and can also be used as medicine. We describe the complete mitochondrial genome of the lichenized fungi Usnea jiangxiensis. It is a circular molecule of 62,531 bp in size, and all genes show the typical gene arrangement conforming to the mold consensus. The mitochondrial genome sequence of U. jiangxiensis and other eight species were used for phylogenetic analysis by the maximum likelihood method. Phylogenetic analysis demonstrated that U. jiangxiensis most closely related to U. ceratina. The complete chloroplast genomes of U. jiangxiensis would be useful for future investigation of genetics, evolution and clinical identification of Usnea species.
Usnea jiangxiensis known as ‘Old Man’s Beard’, which belongs to the genus Usnea within the family Usneaceae (Ascomycota: Parmeliaceae), is mainly distributed in Jiangxi, Shanxi and Gansu Province of China and grows on the bark (Kirk et al. Citation2008). Diagnostic features of the genus include a fruticose thallus with a cortex, medulla, and a cartilaginous central axis, and the presence of usnic acid in the cortex.
The samples of U. jiangxiensis were collected from the forest of the village called Dajing of the Mount Jinggang in Jiangxi Province(Latitude: 26.570583 N, Longitude: 114.138321E),China and 99% same to KX987159.1 in GenBank by rrnL sequence identification. A voucher specimen is deposited at the Key Laboratory of Microbial Biochemistry and Etabolism Engineering of Zhejiang Province with accession number 20190802UJ. Genomic DNA was prepared using CTAB method as described previously (Sun et al. Citation2013). One short sequencing library(ca.300 bp) were constructed and sequenced using illumine novaseq platform. Genome assembly and annotation were performed by CLC Genomies Workbench and Geseq tool (Tillich et al. Citation2017)with default settings. One contig representing the mitochondrial DNA was identified on the basis of extensive sequence similarity to known fungal mitochondrial genomes. The whole genome sequence data reported in this paper have been deposited in the Genome Warehouse in National Genomics Data Center (BIG Data Center Members Citation2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number GWHABKU00000000 that is publicly accessible at https://bigd.big.ac.cn/gwh. The complete mitochondrial genome sequence of U. jiangxiensis is 62,531 bp in length and AT content of 61.50%.
The complete mitochondrial genome contains 14 protein-coding genes (PCGs), 20 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) gene, and one control region (D-loop). All genes show the typical gene arrangement conforming to the Mold mitochondrial consensus (Noack et al. Citation1996). The genome presented here contained a conserved set of 14 protein coding genes (cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, atp6, atp8, and rps3). U. jiangxiensis contained a large intergenic region between LSU and nad2, which is 6.83 kb in size and containing 8 tRNA sequences. 10 introns invade 5 genes including nad1 (two), rps3 (one), atp6 (one), cob (four), nad5 (two).
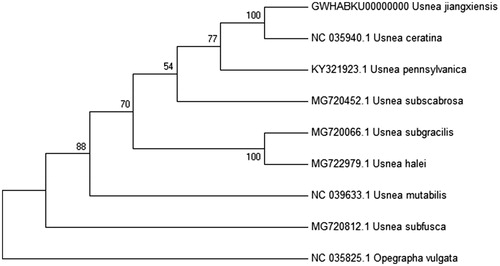
Phylogenetic analysis based on 14 conserved Protein-coding genes using Mega 7.0 (Kumar et al. Citation2016) with ML method confirms U. jiangxiensis as a member of the Lichen order Usnea, which is clustered together with U. ceratina, U. subscabrosa, U. subgracilis, U. halei, U. mutabilis, U. pennsylvanica and U. subfusca (Funk et al. Citation2018). Opegrapha vulgata is set as an outgroup. The phylogenetic tree indicated that U. jiangxiensis was most closely related to U. ceratina in the genus Usnea ().
Figure 1. Phylogenetic analysis of U. jiangxiensis and related species mitochondrial genome. The ML-tree is based on 14 concatenated core mitochondrial proteins respectively from 8 Usnea genomes. Numbers at the nodes are bootstrap values from 2000 replicates. Opegrapha vulgata (NC_035825.1) was set as the outgroup. All other sequences were downloaded from NCBI GenBank. Evolutionary analyses were conducted in MEGA7 (Kumar et al. Citation2016).
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- BIG Data Center Members 2019. Database resources of the big data center in 2019. Nucleic Acids Res. 47:D8–D14.
- Funk ER, Adams AN, Spotten SM, Van Hove RA, Whittington KT, Keepers KG, Pogoda CS, Lendemer JC, Tripp EA, Kane NC, et al. 2018. The complete mitochondrial genomes of five lichenized fungi in the genus Usnea (Ascomycota: Parmeliaceae). Mitochondrial DNA Part B. 3(1):305–308.
- Kirk P, Minter DS. 2008. Dictionary of the Fungi(tenth edition)[M]. CABI Europe-UK.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Noack K, Zardoya R, Meyer A. 1996. The complete mitochondrial DNA sequence of the bichir (Polypterus ornatipinnis), a basal ray-finned fish: ancient establishment of the consensus vertebrate gene order. Genetics. 144(3):1165–1180.
- Sun X, Ruan R, Lin L, Zhu C, Zhang T, Wang M, Li H, Yu D. 2013. Genomewide investigation into DNA elements and ABC transporters involved in imazalil resistance in Penicillium digitatum. FEMS Microbiol Lett. 348(1):11–18.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucl Acids Res. 45(W1):W6–W11.
