Abstract
Catalpa ovata is an ornamental tree species belonging to the Bignoniaceae family and also a valuable wood production plant. In this study, we sequenced the complete chloroplast genome of C. ovata and discussed its phylogenetic relationship. The complete chloroplast genome of C. ovata is 158,279 bp in size, comprising of a large single-copy (LSC, 85,004 bp) and a small single-copy (SSC, 12,675 bp) regions, separated by two inverted repeat regions (IRs, 30,300 bp each). The total GC content was 38.1%. The chloroplast genome contains 131 genes, including 86 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Phylogenetic analysis showed that the complete chloroplast genome sequence of C. ovata can be used as a super DNA barcoding to distinguish it from other plants. This study will provide genetic resources for species identification of C. ovata.
Catalpa Scop. is a perennial tree commonly used for horticulture as garden and street trees. It is generally believed that there are 11 species, five of which are native to China (Wang et al. Citation2016). Catalpa ovata is a pod-bearing tree native to China, which has good ornamental value and is often described as the most beautiful hardy flowering tree (Olsen and Kirkbride Citation2017). Pharmacological studies have shown that catalpa possesses diverse pharmacological activities, including anti-inflammatory and antioxidant (Kil et al. Citation2018; Kim et al. Citation2019). Therefore, the development and utilization of c. ovata has an important economic value. However, catalpa trees received from commercial and noncommercial sources are often misidentified (Olsen and Kirkbride Citation2017). Molecular systematics has become an important method for species identification. Several genes, including the nrDNA ITS (The internal transcribed spacer of ribosomal DNA) and chloroplast ndhF genes, have indicated that genus Catalpa Scop is strongly related to genus Chilopsis D. Don in Bignoniaceae, and C. ovata is more closely related to C. speciosa and C. bignonioides (Li Citation2008). As a useful molecular marker with more genetic information than a single gene, the chloroplast genomes have been widely used in species identification (Fan et al. Citation2019; Yang et al. Citation2019). However, so far, no chloroplast genome of C. ovata has been described. In this paper, the complete chloroplast genome of C. ovata was sequenced and assembled, aiming to provide new genetic information for the germplasm of C. ovata.
The leaf samples of C. ovata were collected from the west campus of Hebei North University in Zhangjiakou city, Hebei province, China (116°12′12″E, 40°0′2″N), The specimen was stored in the herbarium of traditional Chinese medicine, Hebei Northern University (Specimen code HB201909). Total genomic DNA was extracted from fresh leaves using the modified CTAB method described earlier (Uddin et al. Citation2014), The DNA libraries were sequenced by the Illumina pair-end technology and produced a 150 bp pair-end reads. Raw sequence reads were edited using NGS QC Tool kit (Patel and Jain Citation2012). The quality-checked reads were then assembled de novo by SPAdes (Bankevich et al. Citation2012) and the assembled genome was annotated using Plann software (Huang and Cronk Citation2015). Finally, the verified complete chloroplast genome sequence of C. ovata was submitted to GenBank (Accession number MT063116). Phylogenetic position of C. ovata was inferred using the whole plastome sequences, 14 complete chloroplast genomes were aligned by HomBlocks software (Bi et al. Citation2018), and then the multiple sequence alignments were trimmed using Gblock software (Castresana Citation2000). Finally, phylogenetic reconstruction was performed using the maximum-likelihood method implemented in RAxML v8.2.9 software (Stamatakis Citation2014), all the node reliabilities were computed using 1000 bootstrap replicates.
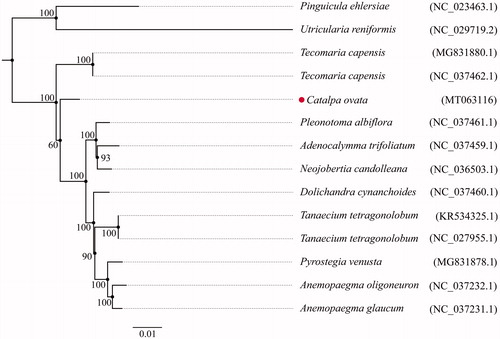
The complete chloroplast genome of C. ovata is 158,279 bp in length, with a typical quadripartite structure including two copies of inverted repeated regions (IRs, 30,300 bp each) that are separated by a large single-copy region (LSC, 85,004 bp) and a small single-copy region (SSC, 12,675 bp). The GC content of the whole chloroplast genome is 38.1%. The plastome contains 131 genes, including 86 protein-coding genes, 8 ribosomal RNA genes, and 37 tRNA genes. The result of the phylogenetic tree showed that the chloroplast genome of C. ovata has enough genetic information to distinguish it from other plants (). This information is crucial for the correct identification of C. ovata and provides valuable genetic resources for the future development of chloroplast derived molecular markers.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Bi G, Mao Y, Xing Q, Cao M. 2018. HomBlocks: A multiple-alignment construction pipeline for organelle phylogenomics based on locally collinear block searching. Genomics. 110(1):18–22.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552.
- Fan J, Li XJ, Li CH, Yang BN. 2019. The complete chloroplast genome and phylogenetic analysis of Begonia pulchrifolia, a near endangered Begoniaceae plant. Mitochondr DNA B. 4(2):2830–2831.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Kil YS, So YK, Choi MJ, Han AR, Jin CH, Seo EK. 2018. Cytoprotective dihydronaphthalenones from the wood of Catalpa ovata. Phytochemistry. 147:14–20.
- Kim HY, Han AR, Kil YS, Seo EK, Jin CH. 2019. Anti-inflammatory effects of catalpalactone isolated from Catalpa ovata in LPS-induced RAW264.7 cells. Molecules. 24(7):1236.
- Li J. 2008. Phylogeny of Catalpa (Bignoniaceae) inferred from sequences of chloroplast ndhF and nuclear ribosomal DNA. J Systemat Evol. 46:341–348.
- Olsen RT, Kirkbride JH. 2017. Taxonomic revision of the genus Catalpa (Bignoniaceae). Brittonia. 69(3):387–421.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Uddin MS, Sun W, He X, Teixeira Da Silva JA, Cheng QX. 2014. An improved method to extract DNA from mango Mangifera indica. Biologia. 69(2):133–138.
- Wang P, Ma Y, Ma L, Li Y, Wang S, Li L, Yang R, Wang Q. 2016. Development and characterization of EST-SSR markers for Catalpa bungei (Bignoniaceae). Appl Plant Sci. 4(4):1500117.
- Yang Z, Huang Y, An W, Zheng X, Huang S, Liang L. 2019. Sequencing and structural analysis of the complete chloroplast genome of the medicinal plant Lycium Chinense Mill. Plants. 8(4):87.