Abstract
Styrax tonkinensis Craib ex Hartwich is a deciduous species of Styracaceae family with beautiful shape, drooping flowers, blooming like snow. Here, we characterized the complete chloroplast (cp) genome of S. tonkinensis using next-generation sequencing method. The circular complete cp genome of S. tonkinensis is 157,934 bp in length, containing a large single-copy (LSC) region of 87,480 bp, and a small single-copy (SSC) region of 18,354 bp. It comprises 134 genes, including 8 rRNA genes, 37 tRNAs genes, and 89 protein-coding genes. The GC content of S. tonkinensis cp genome is 36.92%. The phylogenetic analysis suggests that S. tonkinensis is a sister species to S. obassia in Styracaceae.
Styrax tonkinensis Craib ex Hartwich is an outstanding member of Styracaceae family, mainly distributed in tropical and subtropical lowland areas with elevations of 30–2700 m. It possesses a wide ecological amplitude and strong ability of germination and regeneration. Currently, S. tonkinensis possesses high value for ornamental, timber, oil and medicinal purposes. Its utilization value has been gradually recognized, and it will be widely used in the future (Huang and Grimes Citation2003). But, to date, complete cp genome has not been characterized for S. tonkinensis. In this study, we characterized the complete cp genome sequence of S. tonkinensis (GeneBank accession number: MT075718) based on Illumina pair-end sequencing to provide a valuable complete cp genomic resource.
Total genomic DNA was isolated from the fresh leaves of S. tonkinensis grown in Guantanghe (N32.4544, E118.9546), Luhe District, Nanjing, Jiangsu, China. The voucher specimen was deposited at the herbarium of Nanjing Forestry University (accession number NF2019868). The whole genome sequencing was carried out on Illumina Hiseq platform by Nanjing Genepioneer Biotechnology Inc. (Nanjing, China). The original reading was filtered by CLC Genomics Workbench v9, and the clean reading was assembled into chloroplast genome with SPAdes (Bankevich et al. Citation2012). Finally, CpGAVAS (Liu et al. Citation2012) was used to annotate the gene structure and OGDRAW (Lohse et al. Citation2013) was used to generate the physical map. Based on the neighbor-joining (NJ) method, the phylogenetic tree was deduced by MAFFT (Katoh and Standley Citation2013) and MEGA version 7 (Kumar et al. Citation2016).
The circular genome of S. tonkinensis was 157,934 bp in size and contained two inverted repeat (IRa and IRb) regions of 26,050 bp, which were separated by a large single-copy (LSC) region of 87,480 bp, and a small single-copy (SSC) region of 18,354 bp. A total of 134 genes are encoded, including 89 protein-coding genes (81 PCG species), 37 tRNA genes (30 tRNA species), and 8 rRNA genes (4 rRNA species). Most of the genes occurred in a single copy; however, 8 protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf1, ycf2 and ycf15), 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG and trnV-GAC), and 4 rRNA genes (4.5S, 5S, 16S, and 23S) are totally duplicated. A total of 9 protein-coding genes (atpF, ndhA, ndhB, petB, petD, rpl16, rpoC1, rps16 and rpl2) contained 1 intron while the other 3 genes (clpP, ycf3, rps12) had 2 introns each. The overall GC content of S. tonkinensis genome is 36.92%, and the corresponding values in LSC, SSC, and IR regions are 34.77%, 30.13%, and 42.93%, respectively.
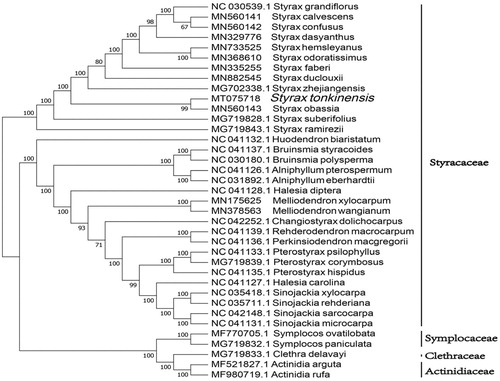
The phylogenetic analysis was conducted based on 32 Styracaceae cp genomes and 3 taxa (Symplocaceae, Actinidiaceae, and Clethraceae) as outgroups with sequenced cp genomes. We found that S. tonkinensis was clustered with other families of Styracaceae with 100% boot-strap values (). In addition, S. tonkinensis was highly supported to be a sister species to Styrax obassia in Styracaceae.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Huang S-M, Grimes JW. 2003. Styracaceae. In: Wu Z-Y, Raven PH, Hong D-Y, editors. Flora of China, Vols. 15 (Styracaceae). Beijing: Science Press; St. Louis: Missouri Botanic Garden Press; p. 258.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability (Article). Mol Biol Evol. 30(4):772–780.
- Kumar S, Stecher G, Tamura K. 2016. Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Liu C, Shi LC, Zhu YJ, Chen HM, Zhang JH, Lin XH, Guan XJ. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. Organellar Genome DRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.