Abstract
We sampled the first mitogenomes from a novel lineage of the Chlorophyceae that is sister to Sphaeropleales. Although the Jenufa perforata (27,180 bp) and Jenufa minuta (41,488 bp) mitogenomes differ markedly in size, gene order and intron content, their repertoires of conserved genes are essentially identical. Their rRNA genes display the same fragmentation patterns as their counterparts in Sphaeropleales. Their tRNA gene complements include two ancestral genes shared with Oedogoniales and Chaetophorales (Chlorophyceae) as well as a derived gene for a tRNA species decoding UGA as tryptophan. This noncanonical genetic code differs from those previously reported for Sphaeropleales.
Mitogenome evolution in the green algal class Chlorophyceae is characterized by the acquisition of a reduced-derived pattern. Representatives of four of the five orders in this class (Chlamydomonadales (Volvocales) + Sphaeropleales clade, Oedogoniales and Chaetophorales) were previously sampled for mitogenome analysis, providing insights into the suite of events that transform this genome (Nedelcu et al. Citation2000; Smith et al. Citation2013; Fucikova et al. Citation2014; Turmel, Bélanger, et al. Citation2020). Gene content underwent severe reduction before the emergence of Chlamydomonadales (12 genes versus 37–42 in other lineages), while modifications of tRNA genes and codon usage led to numerous changes of the genetic code during the evolution of Sphaeropleales (Noutahi et al. Citation2019; Zihala and Elias Citation2019). In contrast to Chlamydomonadales and Sphaeropleales, rRNA genes of Oedogoniales and Chaetophorales underwent no or very little fragmentation and the products of their tRNA genes can decode all codons present in protein-coding genes using the standard genetic code (Turmel, Bélanger, et al. Citation2020). Here, we present the mitogenomes of two species from the genus Jenufa, which represent a novel lineage of the Chlorophyceae that was resolved as sister to Sphaeropleales in plastid phylogenomic trees (Lemieux et al. Citation2015).
Jenufa minuta (CAUP H8102) and Jenufa perforata (CAUP H8101) were obtained from the Culture Collection of Algae at Charles University in Prague and their DNAs were sequenced on the Roche 454 and Illumina MiSeq platforms, respectively, by the Genomic Analysis Platform of Laval University. Given that these sequence data were used to assemble the plastomes, the reader is referred to Lemieux et al. (Citation2015) for detailed description of DNA extraction, sequencing and assembly.
The mitogenomes of J. perforata (27,180-bp; GenBank MN933931) and J. minuta (41,488-bp; GenBank MN933932) differ substantially in size, gene organization and intron content. However, with the exception of an extra tRNA gene [trnW(cca)] present in J. perforata, they possess identical repertoires of conserved genes, which include 13 respiratory protein-coding genes. As in Sphaeropleales, the Jenufa rns and rnl genes occur as two and four separate fragments, respectively. Of the 25 tRNA genes identified in J. perforata, two [trnI(cau) and trnS(uga)] were previously observed only in Oedogoniales and Chaetophorales among Chlorophyceae, while trnW(uca) is missing from all chlorophycean mitogenomes examined thus far. The latter gene, derived from a duplicated copy of trnW(cca) (J. perforata trnW(cca) and trnW(uca) are located side by side), encodes a tRNATrp species that recognizes UGA, normally a stop codon, as well as UGG. All UGA codons found in the Jenufa mitogenomes occur within internal coding sequences of protein-coding genes, corroborating the notion that UGA serves as a sense codon. Among green algae, the noncanonical genetic code reported here was documented for mitochondria originating from Pedinophyceae (Turmel et al. Citation1999) and the prasinophyte Pycnococcus provasolii (Turmel et al. Citation2010).
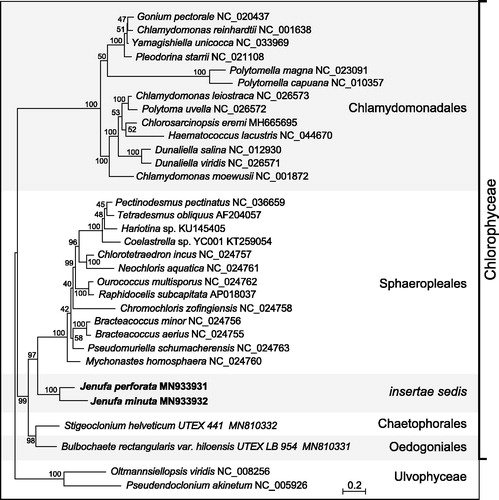
As expected, phylogenetic analysis of 13 mitogenome-encoded proteins from 30 chlorophycean green algae using RAxML v.8.2.3 (Stamatakis Citation2014) recovered the two Jenufa species as a strongly supported clade at a position sister to Sphaeropleales ().
Figure 1. RAxML tree inferred from 13 concatenated mitogenome-encoded proteins of 30 chlorophycean green algae. Oltmannsiellopsis viridis and Pseudendoclonium akinetum (Ulvophyceae) were used as outgroup taxa. The figure shows the best-scoring tree, with bootstrap support values (100 replicates) reported on the nodes. GenBank accession numbers are provided for the mitogenomes of all taxa. The scale bar denotes the estimated number of amino acid substitutions per site. The data set was generated using the predicted protein sequences derived from atp6, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6. Following alignment of the sequences of individual proteins with Muscle v3.7 (Edgar Citation2004), ambiguously aligned regions were removed using TrimAL v1.4 (Capella-Gutierrez et al. Citation2009) with the options block = 6, gt = 0.7, st = 0.005 and sw = 3, and the protein alignments were concatenated using Phyutility v2.2.6 (Smith and Dunn Citation2008). The phylogenetic analysis was carried out under the GTR + Γ4 model.
Disclosure statement
The authors report no conflict of interest. The authors alone are responsible for the content and writing of this article.
Additional information
Funding
References
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797.
- Fucikova K, Lewis PO, Gonzalez-Halphen D, Lewis LA. 2014. Gene arrangement convergence, diverse intron content, and genetic code modifications in mitochondrial genomes of Sphaeropleales (Chlorophyta). Genome Biol E. 6(8):2170–2180.
- Lemieux C, Vincent AT, Labarre A, Otis C, Turmel M. 2015. Chloroplast phylogenomic analysis of chlorophyte green algae identifies a novel lineage sister to the Sphaeropleales (Chlorophyceae). BMC Evol Biol. 15(1):264.
- Nedelcu AM, Lee RW, Lemieux C, Gray MW, Burger G. 2000. The complete mitochondrial DNA sequence of Scenedesmus obliquus reflects an intermediate stage in the evolution of the green algal mitochondrial genome. Genome Res. 10(6):819–831.
- Noutahi E, Calderon V, Blanchette M, El-Mabrouk N, Lang BF. 2019. Rapid genetic code evolution in green algal mitochondrial genomes. Mol Biol E. 36(4):766–783.
- Smith DR, Hamaji T, Olson BJ, Durand PM, Ferris P, Michod RE, Featherston J, Nozaki H, Keeling PJ. 2013. Organelle genome complexity scales positively with organism size in volvocine green algae. Mol Biol E. 30(4):793–797.
- Smith SA, Dunn CW. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Turmel M, Bélanger A-S, Otis C, Lemieux C. 2020. Complete mitogenomes of the chlorophycean green algae Bulbochaete rectangularis var. hiloensis (Oedogoniales) and Stigeoclonium helveticum (Chaetophorales) provide insight into the sequence of events that led to the acquisition of a reduced-derived pattern of evolution in the Chlamydomonadales and Sphaeropleales. Mitochondrial DNA Part B. 5(1):611–613.
- Turmel M, Lemieux C, Burger G, Lang BF, Otis C, Plante I, Gray MW. 1999. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor: two radically different evolutionary patterns within green algae. Plant Cell. 11(9):1717–1730.
- Turmel M, Otis C, Lemieux C. 2010. A deviant genetic code in the reduced mitochondrial genome of the picoplanktonic green alga Pycnococcus provasolii. J Mol Evol. 70(2):203–214.
- Zihala D, Elias M. 2019. Evolution and unprecedented variants of the mitochondrial genetic code in a lineage of green algae. Genome Biol E. 11(10):2992–3007.
