Abstract
Perilla frutescens is well known for its high α-linolenic acid (ALA) accumulation and medicinal values in the world. In this study, the complete chloroplast genome of P. frutescens was reported. The chloroplast genome length of P. frutescens was 152,602 bp, which was circular in shape with a characteristic quadripartite structure. It contained a large single-copy (LSC) region of 83,706 bp, a small single-copy (SSC) region of 17,546 bp and separated by two inverted repeat (IR) regions of 25,675 bp. The overall nucleotide content of the chloroplast genome was 38.1% G + C content. This chloroplast genome contained 139 genes, which included 94 protein-coding genes (PCGs), 37 transfer RNA (tRNAs), and 8 ribosome RNA (rRNAs). Phylogenetic and evolutionary relationship have shown that P. frutescens was closely related to P. setoyensis in the family Lamiaceae using the maximum-likelihood (ML) method.
Perilla frutescens (Zi-Su in Chinese) is a self-pollinating plant and widely distributed in East Asian countries and considered as a food supply and natural medicine resource (Yu et al. Citation2017). It belongs to the family Lamiaceae, which has been used as an herb medicine in China (Lee and Ohnishi Citation2003). Perilla frutescens is used in herbal medicine to treat a wide variety of ailments, as well as in cooking as a garnish and as a possible antidote to food poisoning, which has shown antioxidant, antiallergic, anti-inflammatory, antidepressant, anorexigenic, and tumor-preventing properties. However, there are limited clinical data to support the use of Perilla (Liao et al. Citation2018). In this paper, the chloroplast genome of P. frutescens has been presented provides valuable genomic resources of the plant chloroplast genome, which will extend our information of TCM business in the world.
The fresh sample of P. frutescens was collected from the herb market near Zhejiang Chinese Medical University (Hangzhou, Zhejiang, China, 119.89°E, 30.09°N). The chloroplast DNA of P. frutescens was extracted from the fresh blades by the modified CTAB method, which was stored at the Zhejiang Chinese Medical University (No. ZJCMU-004). The chloroplast DNA was purified and sequenced. For quality control, the low-quality reads were removed by the FastQC software (Andrews Citation2015). The chloroplast genome of P. frutescens was assembled and annotated by the MitoZ software (Meng et al. Citation2019). The physical map of the chloroplast genome of P. frutescens was drawn by the OGDRAW software (Greiner et al. Citation2019). The sequence information of chloroplast genome had been submitted to NCBI GenBank with the accession No. MN04467322.
The chloroplast genome length of P. frutescens was 152,602 bp, which circular in shape with a characteristic quadripartite structure. It contained a large single-copy (LSC) region of 83,706 bp, a small single-copy (SSC) region of 17,546 bp, and separated by two inverted repeat (IR) regions of 25,675 bp. The chloroplast genome of P. frutescens contained 139 genes, which included 94 protein-coding genes (PCGs), 37 transfer RNA genes (tRNAs), and 8 ribosomal RNA genes (rRNAs). That total of 17 genes with 8 PCGs species, 7 tRNA genes species and 4 rRNA genes species were found duplicated in one of the IR regions. The overall nucleotide content of the chloroplast genome: 30.7% of A (Adenine, 46,857 bp), 31.5% of T (Thymine, 48,025 bp), 19.2% of C (Cytosine, 29,339 bp), 18.9% of G (Guanine, 28,381 bp), and 38.1% G + C content.
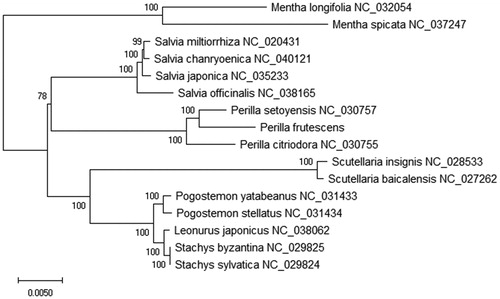
To further investigate the phylogenetic and evolutionary relationship of Perilla frutescens, we constructed the phylogenetic tree based on 16 plant chloroplast genomes. The MEGA X software (Kumar et al. Citation2018) was used for the phylogenetic tree analysis based on the bootstrap values of 2000 replicates for all the nodes and ML method was used to construct the tree. The ML phylogenetic tree was edited using the iTOL version 4.0 (Letunic and Bork Citation2016). Phylogenetic and evolutionary relationship showed that P. frutescens was closely related to P. setoyensis in the family Lamiaceae (). This study can provide a platform to elucidate the use of P. frutescens in TCM.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Andrews S. 2015. FastQC: a quality control tool for high throughput sequence data. [accessed 2020 Mar 14]. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549.
- Lee JK, Ohnishi O. 2003. Genetic relationships among cultivated types of Perilla frutescens and their weedy types in East Asia revealed by AFLP markers. Genet Resour Crop Evol. 50(1):65–74.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v4: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245.
- Liao B, Hao Y, Lu J, Bai H, Guan L, Zhang T. 2018. Transcriptomic analysis of Perilla frutescens seed to insight into the biosynthesis and metabolic of unsaturated fatty acids. BMC Genom. 19(1):213.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63–e63.
- Yu H, Qiu JF, Ma L, Hu YJ, Li P, Wan JB. 2017. Phytochemical and phytopharmacological review of Perilla frutescens L. (Labiatae), a traditional edible-medicinal herb in China. Food Chem Toxicol. 108(Pt B):375–391.