Abstract
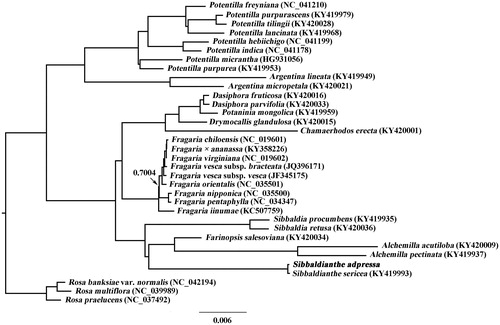
The complete chloroplast genome of Sibbaldianthe adpressa was reported in this study. The chloroplast genome of S. adpressa was a circular form of 155,021 bp in length, consisting of a pair of inverted repeats (IRa and IRb) of 26,028 bp separated by a large single copy (LSC) region of 84,712 bp and a small single copy (SSC) region of 18,253 bp. The genome contained a set of 129 genes, comprising 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes. Phylogenetic analysis indicated that S. adpressa was sister to S. sericea.
Sibbaldianthe adpressa (Bunge) Juz. (synonym: Sibbaldia adpressa Bunge), belongs to the family Rosaceae Juss., subfamily Rosoideae (Juss.) Arn., tribe Potentilleae Sweet. Its native range is Central Asia to Mongolia and North Nepal according to Plants of the World online maintained by the Royal Botanic Gardens, Kew, UK (http://powo.science.kew.org). The complete chloroplast (cp) genome of S. adpressa was reported herein, which is helpful for further studies on its taxonomy and population genetics.
Fresh leaves of S. adpressa were collected from Chahar Right Back Banner, Nei Mongol, China. The voucher specimen (no. Li QQ 20160430005) was deposited in the herbarium of Inner Mongolia Normal University (NMTC). Total genomic DNA was extracted using the CTAB method (Doyle and Doyle Citation1987). The library with an insert size of 300 bp fragments was prepared and then sequenced using an Illumina HiSeq 2000 Sequencing System in Novogene (Nanjing, China). Illumina paired-end sequencing generated 39,067,938-bp raw reads after adapters were removed. The raw reads were used to assemble the cp genome in NOVOPlasty (Dierckxsens et al. Citation2017) with ribulose-1,5-bisphosphate carboxylase/oxygenase (rbcL) gene from Sibbaldianthe sericea (GenBank accession no. KY419993) as the seed. Chloroplast genome annotation was performed by transferring annotations in Geneious Prime (Kearse et al. Citation2012), with complete cp genome of Farinopsis salesoviana (Steph.) Chrtek et Soják (GenBank accession no. MT017928) as the reference. The positions of start and stop codons and intron/exon boundaries were manually checked. The annotated complete cp genome of S. adpressa was deposited in GenBank with accession no. MT114191. The complete cp genome of S. adpressa was circular in form with a size of 155,021 bp in length and had a typical quadripartite structure consisting of a pair of inverted repeats (IRa and IRb: 26,028 bp) separated by a large single-copy (LSC: 84,712 bp) and a small single-copy (SSC: 18,253 bp) regions. The overall GC content was 37.2%. The cp genome contained a set of 129 genes, comprising 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes.
To ascertain the phylogenetic position of S. adpressa, the cp genome sequences of 30 Potentilleae taxa plus three Rosa taxa were downloaded from GenBank. All the cp genome sequences were aligned by MAFFT version 7.450 (Katoh and Standley Citation2013) and then trimmed properly using trimAL version 1.4 (Capella-Gutiérrez et al. Citation2009). Phylogenetic analysis was conducted by employing Bayesian inference (BI) using the MrBayes version 3.2.2 (Ronquist et al. Citation2012). Prior to the Bayesian analysis, the GTR + I + G model under the Akaike infomation criterion (AIC, Posada and Buckley Citation2004) was selected as a best-fit model using PartitionFinder2 (Lanfear et al. Citation2016). Four independent Markov chain Monte Carlo (MCMC) algorithms were run for 6,000,000 generations, starting from random trees and sampling trees every 100 generations. The first 15,000 trees were considered as the burn-in and were discarded. A 50% majority-rule consensus tree was produced. The phylogenetic tree indicated that S. adpressa was sister to S. sericea ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Capella-Gutiérrez S, Silla‐Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochem Bull. 19(1):11–15.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones‐Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2016. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Posada D, Buckley TR. 2004. Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 53(5):793–808.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542.