Abstract
Malva verticillata, which belongs to the family Malvaceae, is an herb with a wide ecological range and considerable economic and ecological value. herein this study, we determined the complete chloroplast (cp) genome of M. verticillata using Illumina sequencing. The total length of the cpDNA genome was 158,408 bp, including two copies of inverted repeats (IRs, 50,214 bp), a large single copy (LSC, 87,085 bp), and a small single copy (SSC, 21,109 bp). A total of 130 genes were annotated, including 36 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes, and 86 messenger RNA (mRNA) genes. Phylogenetic analysis showed that M. verticillata is closely related to Abelmoschus esculentus, and Hibiscus syriacus among Malvaceae species.
Malva verticillata (Malvaceae, Malva), an annual herb with a wide ecological range, is mainly found in India, central Asia, Siberia, Japan, Mongolia, Europe and China (Feng Citation1984). Malva verticillata has been planted as vegetable, medicinal plant, and forage plant in China because of its good taste and various medicinal components (Bao et al. Citation2018). Besides, it is well adapted to the drought and saline-alkaline land (Wang et al. Citation2002; Bao et al. Citation2018), which possesses its economic and ecological value. It is important to study its genetic diversity, protect its wild germplasm resources, make genetic improvements, and cultivate further exploitation and utilization. However, there is currently no theoretical basis for these necessary work. As chloroplast (cp) genes are widely used in studies of genetic diversity and phylogeny (Fu et al. Citation2016), we determined the complete cp genome of M. verticillata based on next-generation sequencing data, which we expect will help future studies.
Total genomic DNA was extracted from fresh and clean leaves of M. verticillata sampled from Xining City (N36°35′10.09″, E101°49′18.28″) in China’s Qinghai Province with a modified CTAB method (Doyle and Doyle Citation1987). The specimen was stored at Herbarium of the Northwest Institute of Plateau Biology (HNWP, Wang2020001), Northwest Institute of Plateau Biology, Chinese Academy of Sciences. Genome sequencing was performed using the Illumina Novaseq Platform (Illumina, San Diego, CA) with an Illumina pair-end at Genepioneer Biotechnologies Inc., Nanjing, China. Raw reads were filtered using OpenGene/fastp (https://github.com/OpenGene/fastp) (Chen et al. Citation2018). The clean reads were assembled with core module from SPAdes v3.10.1 (http://cab.spbu.ru/software/spades/) (Anton et al. Citation2012) and aligned to the company’s (Genepioneer Biotechnologies Inc., Nanjing, China) own cp genome database. The online tools Blast v2.2.25 (https://blast.ncbi.nlm.nih.gov/Blast.cgi), Hmmer v3.1b2 (http://www.hmmer.org/), and Aragorn v1.2.38 (http://130.235.244.92/ARAGORN/) were used to annotate messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA) sequences, respectively. The complete cp genome sequence of M. verticillata was first deposited in NCBI Genbank (accession number: MT083899). OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) was performed to produce the complete cp genome map. The phylogenetic tree was generated based on M. verticillata and other Malvaceae species using RAxML software with a GTR model, hill climbing method, and bootstrap set to 1000 (Stamatakis Citation2014).
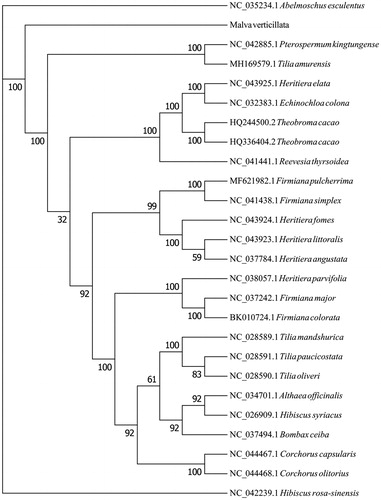
The new sequence yielded approximately 7.9 GB of clean data. The assembled M. verticillata cp genome is a typical circular DNA 158,408 bp in length, including two copies of inverted repeats (IRs, 50,214 bp), a large single copy (LSC, 87,085 bp), and a small single copy (SSC, 21,109 bp). Overall GC content was 37.12%, and those in LSC, SSC, and IR regions were 34.94%, 32.11%, and 43.01%, respectively. A total of 130 genes were annotated, including 36 tRNA genes, 8 rRNA genes, and 86 mRNA genes. The phylogenetic results () showed that Malvaceae species are divided into three obvious lineages, and M. verticillata is most similar to Abelmoschus esculentus, followed by Hibiscus syriacus.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Anton B, Sergey N, Dmitry A, Alexey AG, Mikhail D, Alexander SK, Valery ML, Sergey IN, Son P, Andrey DP, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell. J Comput Biol. 19(5):455–477.
- Bao LM, Bao XH, Li P, Wang XL, Ao W. 2018. Chemical profiling of Malva verticillata L. by UPLC-Q-TOF-MSE and their antioxidant activity in vitro. J Pharmaceut Biomed. 150:420–426.
- Chen SF, Zhou YQ, Chen YR, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890.
- Doyle JJ, Doyle JL. 1987. A Rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Feng TM. 1984. Flora of China, Vol 49. Beijing: Science Press.
- Fu PC, Zhang YZ, Geng HM, Chen SL. 2016. The complete chloroplast genome sequence of Gentiana lawrencei var. farreri (Gentianaceae) and comparative analysis with its congeneric species. PeerJ. 4:e2540.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Wang GX, Wang XY, Duan YJ. 2002. Study on halophytic structure of vegetative organs of Malva verticillata L. J Zhaowuda Mongolian Teach Coll. 23(3):38–39.