Abstract
Dendrocalamus is an economically important woody bamboo genus. In this study, the complete chloroplast genome of D. yunnanicus was determined from Illumina HiSeq pair-end sequencing data. The genome was 139,432 bp in length, including a large single-copy region (LSC) of 82,964 bp and a small single-copy region (SSC) of 12,878 bp, and two inverted repeat regions (IR) of 21,795 bp. There were 132 genes predicted, containing 85 protein-coding genes, 8 ribosomal RNA genes, and 39 transfer RNA genes.
Dendrocalamus Nees (Poaceae; Bambusoideae; Bambuseae), an economically important woody bamboo genus described in 1835 based on D. strictus (Roxb.) Nees., consists of approximately 52 species collectively distributed in the tropical and subtropical regions of Asia (Ohrnberger Citation1999; Li and Stapleton Citation2006). The challenges of limitation between Dendrocalamus and Bambusa remain in their flowers being rare. In this study, we first reported the complete chloroplast genome of D. yunnanicus.
The total genomic DNA was extracted from fresh leaves issue of D. yunnanicus using a modified CTAB method (Doyle and Doyle Citation1987). The sample was acquired from Cangshan district, Fujian, China (26°38′N, 119°27′E), and the voucher specimen was deposited at the Herbarium of Fujian Agriculture and Forestry University (specimen code HTY005). After DNA extraction, library constructing, sequencing, and data filtering were performed on an Illumina Hiseq 2500 platform, generating ∼20 Gb of sequence data. The chloroplast genome was assembled with the program NOVOPlasty (Dierckxsens et al. Citation2017) with the complete chloroplast genome of its close relative D. latiflorus (GenBank accession NC_013088) as the reference. The assembled chloroplast genome was annotated using Plann (Huang and Cronk Citation2015), and the annotation was corrected using Geneious (Kearse et al. Citation2012). The annotation map of the new chloroplast genome was drawn using OGDRAW (Lohse et al. Citation2013).
The complete D. yunnanicus (GenBank accession MN782326) plastid genome is a quadripartite structure with the length of 139,432 bp, contains a large single-copy region (LSC) of 82,964 bp, a small single-copy (SSC) region of 12,878 bp, and two inverted repeat (IR) regions of 21,795 bp. The new sequences possess total 132 genes, including 85 protein-coding genes, 8 rRNA genes, and 39 tRNA genes. The overall GC content of the chloroplast genome was 38.92%, while the corresponding values of the LSC, SSC, and IR regions are 37.03%, 33.19%, and 44.21%, respectively.
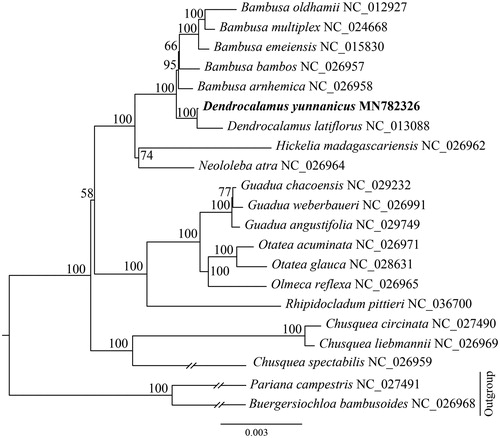
To investigate the phylogenetic position of D. yunnanicus, 20 representative species of Bambuseae were downloaded from the NCBI GenBank database for phylogenetic analysis. The sequences were aligned using MAFFT v7.307(Katoh and Standley Citation2013), and phylogenetic tree was constructed by RAxML (Stamatakis Citation2014). The ML tree showed that D. yunnanicus is sister to D. latiflorus with strong support ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Huang DI, Cronk Q. 2015. Plann: a command-line application for annotating plastome sequences. Appl Plant Sci. 3(8):1500026.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integratreed and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Li DZ, Stapleton C, et al. 2006. Dendrocalamus genus. In: Wu CY, editors. Flora of China 22. Beijing: Science Press; St. Louis: Missouri Botanical Garden Press; p. 39–46.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW—a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(W1):W575–W581.
- Ohrnberger D. 1999. The bamboos of the world: annotated nomenclature and literature of the species and higher and lower taxa. Amsterdam, New York, Oxford, Tokyo: Elsevier Science B.V.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.