Abstract
The complete mitogenome of Mazocraeoides gonialosae parasitized on the gills of dotted gizzard shad (Konosirus punctatus) was obtained. The entire circular mitogenome size of M. gonialosae was 15,106 bp, containing 12 protein-coding genes (PCGs; lacking atp8 gene), 2 rRNA genes, 22 tRNA genes, 1 repeat region, and several non-coding regions. The arrangement of the 12 PCGs was identical with those of other polyophistocotylean monogenean mitogenomes available in GenBank. The phylogenetic tree based on the concatenated 12 PCGs revealed that M. gonialosae belonged to the mazocraeid clade, together with Neomazocraes dorosomatis.
Mazocraeoides gonialosae (Platyhelminthes: Monogenea) belongs to family Mazocraeidae, originally described from the gills of Ganges River gizzard shad (Gonialosa manmina) from India (Tripathi Citation1959). It is also known to parasitize on the gills of dotted gizzard shad (Konosirus punctatus) and Japanese gizzard shad (Nematalosa japonica), distributed along the coast of China (Li et al. Citation2011). Of these hosts, dotted gizzard shad is one of the commercially important fish species in Korea. In this study, we obtained the complete mitochondrial (mt) genome sequences of M. gonialosae parasitizing on the gills of dotted gizzard shad.
The host fish were bought from local fisherman in Jumunjin, Korea (37.53 N, 128.49E) and M. gonialosae were collected from their gill lamellae. Identification was based on the morphological characters (Tripathi Citation1959) and the published mt cox1 gene sequence (Li et al. Citation2011). The slide specimen (specimen number MABIK2019IV0923) was deposited in the National Marine Biodiversity Institute of Korea. DNA was sequenced by MGISEQ-2000 and mt genome assembly was conducted with MITOS (Bernt et al. Citation2013) and tRNAScan (Chan and Lowe Citation2019). Annotation was conducted by BLAST against Neomazocraes dorosomatis (GenBank accession number: JQ038229.1), and phylogenetic relationships between M. gonialosae and other monogeneans were estimated on the basis of the concatenated amino acid sequences of protein-coding genes (PCGs) from selected 24 mongenean mt genomes available in the GenBank. Two Tricladida species (Crenobia alpine and Obama sp.) were used as outgroups.
The complete circular mt genome of M. gonialosae was 15,106 bp in size (GenBank accession number: MT015514), having 12 PCGs (lacking atp8), 22 tRNA genes, 2 rRNA genes, 1 repeat sequence, and several non-coding regions. The nucleotide composition was calculated as 32.64% A, 16.87% G, 9.74% C, and 40.75% T. All the genes were transcribed from the same strand.
The total length of 12 PCGs was 9900 bp, occupying 65.5% of the full mt genome. This is the lowest percentage of total PCGs ratio in the published polyophistocotylean monogenean mt genome. The arrangement of the 12 PCGs of M. gonialosae was identical to those of all the other polyopisthocotylean monogenean mitogenomes in GenBank: cox3-nad6-nad5-cytb-nad4l-nad4-atp6-nad2-nad1-nad3-cox1-cox2. ATG was the start codon for all the PCGs, except for nad1, nad3, and cox3 (GTG as the start codon). Interestingly, ATT was used as the start codon for nad2 gene. TAA was the stop codon for 8 PCGs (nad4l, nad4, atp6, nad2, nad1, nad3, cox1, cox2), while TAG for the rest of them. The size of the 22 tRNA genes was 1498 bp in total, ranging from 61 bp (trnS1) to 72 bp (trnK, trnN). Twenty-one of them had conventional secondary structure, except for trnS1 (lacking DHU arm). The arrangement of tRNA gene was as follows: trnY-trnC-trnK-trnL1-trnS2-trnL2-trnR-trnE-trnQ-trnF-trnD-trnV-trnA-trnN-trnP-trnI-trnS1-trnW-trnG-trnT-trnM-trnH. The size of rrnL and rrnS were 988 bp and 752 bp, respectively. They were adjacent to non-coding regions next to trnT (upstream) and cox2 (downstream), and separated by a short non-coding region (10 bp). There were several non-coding regions (941 bp in total) and one repeat sequence (1039 bp) located between trnK and a non-coding region.
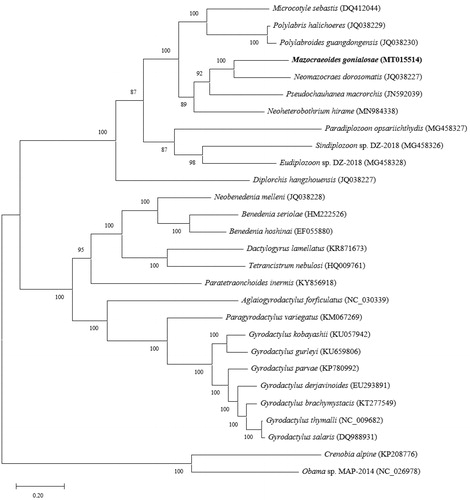
The phylogenetic tree by MEGA X (Kumar et al. Citation2018) revealed that M. gonialosae firmly clustered with N. dorosomatis. These two mazocraeid monogeneans belonged to the polyophistocotylean clade, together with other microcotylid, chauhaneid, diclidophorid and diplozoid monogeneans PCGs available in the GenBank (Park et al. Citation2007; Zhang et al. Citation2011, Citation2012, Citation2018) ().
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Kumar S, et al. 2018.
- Li M, Shi S-F, Brown CL, Yang T-B. 2011. Phylogeographical pattern of Mazocraeoides gonialosae (Monogenea, Mazocraeidae) on the dotted gizzard shad, Konosirus punctatus, along the coast of China. Int J Parasitol. 41(12):1263–1272.
- Park JK, Kim KH, Kang S, Kim W, Eom KS, Littlewood D. 2007. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes). BMC Evol Biol. 7(1):11.
- Tamura K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol E. 35:1547–1549.
- Tripathi YR. 1959. Monogenetic trematodes from fishes of India. Indian J Helminthol. 9:1–149.
- Zhang J, Wu X, Xie M, Xu X, Li A. 2011. The mitochondrial genome of Polylabris halichoeres (Monogenea: Microcotylidae). Mitochondrial DNA. 22(1–2):3–5.
- Zhang J, Wu X, Xie M, Li A. 2012. The complete mitochondrial genome of Pseudochauhanea macrorchis (Monogenea: Chauhaneidae) revealed a highly repetitive region and a gene arrangement hot spot in Polyophistocotylea. Mol Biol Rep. 39(8):8115–8125.
- Zhang D, Zou H, Wu SG, Li M, Jakovlic I, Zhang J, Chen R, Li WX, Wang GT. 2018. Three new Diplozoidae mitogenomes expose unusual compositional biases within the monogenea class: implications for phylogenetic studies. BMC Evol Biol. 18(1):133.