Abstract
Dracocephalum moldavica belongs to the genus Dracocephalum of the Labiatae family. It is a good source of essential oils, and is wildly used in folk medicine. In this study, the complete chloroplast genome of the D. moldavica was sequenced, assembled, and annotated, which contains 125 unique genes, including 88 protein-coding genes, 29 tRNA genes, and 8 rRNA genes. A maximum-likelihood phylogenetic tree based on 13 complete chloroplast genomes revealed that D. moldavica is closely related to D. palmatum. The data could be used for variety identification, species conservation, and cultivation.
The Dracocephalum moldavica L. belongs to genus Dracocephalum of the family Labiatae and mainly distributed within 200–2700 m of dry hills, stony riverbanks, and valleys in Europe and northern Asia (http://www.iplant.cn). It is a good source of protein, essential oil (Wojtowicz et al. Citation2017) and linolenic acid (59.4% in seeds) (Domokos et al. Citation1994). Several studies have shown that the D. moldavica as a herbal drug can be used to treat stomach, liver disorders, headaches, coronary heart disorders, and hypertension (Yang et al. Citation2014).
To date (2/10/2020), more than 30 species belonging to 15 different genus of Labiatae chloroplast genome have been sequenced and released in the National Center for Biotechnology Information (NCBI). However, the chloroplast genome of the D. moldavica has not been reported. In this study, we first report the complete chloroplast genomes of D. moldavica based on Illumina Hiseq pair-end sequencing data.
Fresh and clean leave materials of D. moldavica were collected from Yangling city, Shaanxi Province (34°9′9″N, 108°50′29″E), frozen and preserved at Northwest A&F University. The specimen (No. 61040319082005LY) was deposited in the herbarium of Shaanxi University of Chinese Medicine. Total genomic DNA was extracted with the modified CTAB method (Stefanova et al. Citation2013). Genome sequencing was performed using HiSeqX at Biomarker Technologies Corporation. Low-quality sequences were filtered by NGSQC Toolkit with Q30 (base Phred quality score of ≥30) (Patel and Jain Citation2012). Total high-quality reads were mapped to reference (D. palmatum chloroplast genome: NC_031874) using Bowtie2 (Langmead and Salzberg Citation2012) and the mapped reads were extracted and assembled by MIRA (Chevreux et al. Citation2004) and SPAdes (Bankevich et al. Citation2012). A total of 199,513 reads have been assembled with an average coverage of 200.0×. The assembled chloroplast genome was annotated and corrected using DOGMA (http://dogma.ccbb.utexas.edu/) (Greiner et al. Citation2019)and Geneious (Kearse et al. Citation2012), and deposited into GenBank(accession No. MT104121).
The complete chloroplast genome of D. moldavica is 150,124 bp in length, containing a large single copy (LSC: 82,169 bp), two inverted repeats (IRa and IRb: 25,358 bp), and a small single copy (SSC: 17,239 bp). 125 genes were annotated in total, including 88 protein-coding genes, 29 tRNA genes, and 8 rRNA genes. The GC content of the complete genome is 37.81%.
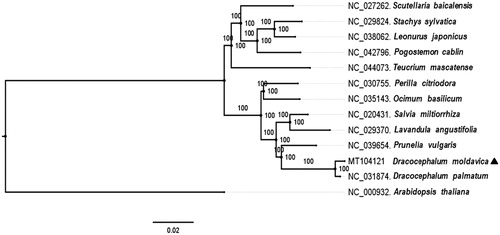
To investigate the D. moldavica taxonomic status, a total of 13 complete chloroplast genome including the D. moldavica, 11 other Labiatae species and Arabidopsis were multiple aligned by MAFFT 7.450 (Katoh et al. Citation2002). Subsequently, a maximum likelihood phylogenetic tree was generated by RAxML v7.2.8 (Stamatakis Citation2006) with 1000 bootstrap replicates (). The results showed that D. moldavica was closely related to the same genus species D. palmatum. Sequences of the complete chloroplast genome of D. moldavica would lay foundation for Chinese medicinal material identification, species conservation, and cultivation.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477.
- Chevreux B, Pfisterer T, Drescher B, Driesel AJ, Muller WEG, Wetter T, Suhai S. 2004. Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14(6):1147–1159.
- Domokos J, Peredi J, Halasz-Zelnik K. 1994. Characterization of seed oils of dragonhead (Dracocephalum moldavica L.) and catnip (Nepeta cataria var. citriodora Balb.). Ind Crops Prod. 3(1–2):91–94.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64.
- Katoh K, Misawa K, Kuma K-i, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690.
- Stefanova P, Taseva M, Georgieva T, Gotcheva V, Angelov A. 2013. A modified CTAB method for DNA extraction from soybean and meat products. Biotechnology Biotechnological Equipment. 27(3):3803–3810.
- Wojtowicz A, Oniszczuk A, Oniszczuk T, Kocira S, Wojtunik K, Mitrus M, Kocira A, Widelski J, Skalicka-Wozniak K. 2017. Application of Moldavian dragonhead (Dracocephalum moldavica L.) leaves addition as a functional component of nutritionally valuable corn snacks. J Food Sci Technol. 54(10):3218–3229.
- Yang L-N, Xing J-G, He C-H, Wu T. 2014. The phenolic compounds from Dracocephalum moldavica L. Biochem Syst Ecol. 54(2014):19–22.