Abstract
The mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae) (GenBank No. MT107158) was a circular molecule of 16,693 bp in length, including 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 non-coding region (control region). The overall base composition of the H-strand of T. roborowskii is 30.6% A, 25.6% T, 29.7% C, 14.1% G, respectively. Phylogenetic analyses showed that T. roborowskii (MT107158) was a sister clade to T. keyserlingii whereas Teratoscincus roborowskii (KP115216) was a close sister clade of T. keyserlingii and T. roborowskii (MT107158).
The genus Teratoscincus (Squamata: Gekkonidae) are endemic species in central and southwest Asia (Macey et al. Citation1999). The species of Gekkonidae are more than nine hundreds while 27 mitogenomes of Gekkonidae were published (Macey et al. Citation2005; Fujita et al. Citation2007; Yan et al. Citation2009; Li et al. Citation2013; Kumazawa et al. Citation2014; Yan, Tian, Lv et al. Citation2014; Yan, Tian, Zhou et al. Citation2014; Kim et al. Citation2016; Li et al. Citation2016; Starostová and Musilová Citation2016; Yan et al. Citation2016; Areesirisuk et al. Citation2018). The phylogenetic relationship among Gekkonidae was controversial in morphological and molecular aspects (Zhou et al. Citation2006; Qin et al. Citation2011; Li et al. Citation2013; Hao et al. Citation2016). To clarify the phylogeny relationships of Gekkonidae, we sequenced the complete mitogenome of T. roborowskii (Squamata: Gekkonidae).
Samples of T. roborowskii were collected in Turpan Prefecture (42°51′33″N, 89°11′29″E), Xinjiang Uygur Autonomous Region, China. The sample (XJTLF20190715) was identified by YP Zhang and stored at −40 °C in the Animal Specimen Museum, College of Life and Environmental Science, Wenzhou University, China. The total genomic DNA was extracted from tail muscle of the sample using Ezup Column Animal Genomic DNA Purification Kit (Sangon Biotech Company, Shanghai, China) and stored in the Zhang’s laboratory. Universal and specific primers for PCR amplification were designed according to Macey et al. (Citation2005). The mitogenome was deposited in GenBank with the accession number MT107158.
The complete mitogenome of T. roborowskii was typical circular DNA molecule of 16,693 bp in length, including 13 protein-coding genes (PCGs), 2 rRNA genes, 22 tRNA genes, and 1 non-coding region (control region). The gene order was similar to the known mitogenomes of gekkonids (Böhme et al. Citation2007; Qin et al. Citation2011; Li et al. Citation2013; Yan et al. Citation2016). Nine PCGs used ATN (N represents A, T, C, G) as the initiation codon, whereas COX1, ATP8, ND4, and ND5 were initiated by GTG. The COX2, COX3, ND4 genes used T as the termination codon and the other PCGs ended with TAA or TAG. The overall base compositions of the H-strand of T. roborowskii was 30.6% A, 25.6% T, 29.7% C, and 14.1% G. The overall AT and GC skew was 0.088 and −0.354, respectively.
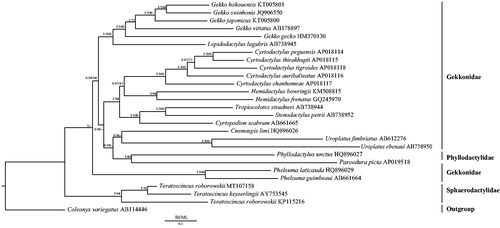
In order to clarify the phylogenetic relationships of gekkonids, 27 sequences of the complete (or nearly complete) mitogenomes were obtained from the GenBank database. Bayesian inference (BI) and maximum likelihood (ML) trees were constructed using the 13 PCGs (). Coleonyx variegatus (Hao et al. Citation2016) was used as the outgroup. BI and ML trees were analyzed by MrBayes 3.1.2 (Huelsenbeck and Ronquist Citation2001) and RAxML 8.2.0 (Stamatakis Citation2014), respectively. Phylogenetic analyses showed that T. roborowskii (MT107158) was a sister clade to T. keyserlingii not T. roborowskii (KP115216). The paraphyly of Gekkonidae was strongly supported in BI and ML analyses as well as the results of Pyron et al. (Citation2013) and Hao et al. (Citation2016) because Phyllodactylus unctus (Phyllodactylidae) was clustered into the clade of Gekkonidae.
Figure 1. Phylogenetic tree of the relationships among 27 species of gekkonids including Teratoscincus roborowskii (MT107158) was based on the nucleotide dataset of the 13 protein-coding genes. Coleonyx variegatus (AB114446) was used as the outgroup. The numbers showed between branches indicate the posteriori probabilities from Bayesian inference (BI) (left) and bootstrap percentages from maximum likelihood (ML) (right). The GenBank accession numbers of all species are shown in the figure.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Areesirisuk P, Muangmai N, Kunya K, Singchat W, Sillapaprayoon S, Lapbenjakul S, Thapana W, Kantachumpoo A, Baicharoen S, Rerkamnuaychoke B, et al. 2018. Characterization of five complete Cyrtodactylus mitogenome structures reveals low structural diversity and conservation of repeated sequences in the lineage. PeerJ. 6:e6121.
- Böhme MU, Fritzsch G, Tippmann A, Schlegel M, Berendonk TU. 2007. The complete mitochondrial genome of the green lizard Lacerta viridis viridis (Reptilia: Lacertidae) and its phylogenetic position within squamate reptiles. Gene. 394(1–2):69–77.
- Fujita MK, Boore JL, Moritz C. 2007. Multiple origins and rapid evolution of duplicated mitochondrial genes in Parthenogenetic geckos (Heteronotia binoei; Squamata, Gekkonidae). Mol Bio Evol. 24(12):2775–2786.
- Hao SL, Ping J, Zhang YP. 2016. Complete mitochondrial genome of Gekko chinensis (Squamata, Gekkonidae). Mitochondrial DNA Part A. 27(6):4226–4227.
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 17(8):754–755.
- Kim IH, Park J, Cheon KS, Lee HJ, Kim JK, Park D. 2016. Complete mitochondrial genome of Schlegel’s Japanese gecko Gekko japonicus (Squamata: Gekkonidae). Mitochondrial DNA Part A. 27(5):3684–3686.
- Kumazawa Y, Miura S, Yamada C, Hashiguchi Y. 2014. Gene rearrangements in gekkonid mitochondrial genomes with shuffling, loss, and reassignment of tRNA genes. BMC Genomics. 15(1):930.
- Li HM, She Y, Hou LX, Zhang Y, Guo DN, Qin XM. 2016. The complete mitochondrial genome of Teratoscincus roborowskii (Squamata: Gekkonidae). Mitochondrial DNA Part A. 27(3):1916–1917.
- Li HM, Zeng DL, Guan QX, Qin PS, Qin XM. 2013. Complete mitochondrial genome of Gekko swinhonis (Squamata, Gekkonidae). Mitochondrial DNA Part A. 24(2):86–88.
- Macey JR, Fong JJ, Kuehl JV, Shafiei S, Ananjeva NB, Papenfuss TJ, Boore JL. 2005. The complete mitochondrial genome of a gecko and the phylogenetic position of the Middle Eastern Teratoscincus keyserlingii. Mol Phylogenet Evol. 36(1):188–193.
- Macey JR, Wang YZ, Ananjeva NB, Larson A, Papenfuss TJ. 1999. Vicariant patterns of fragmentation among gekkonid lizards of the genus Teratoscincus produced by the Indian collision: a molecular phylogenetic perspective and an area cladogram for Central Asia. Mol Phylogenet Evol. 12(3):320–332.
- Pyron RA, Burbrink FT, Wiens JJ. 2013. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol Biol. 13(1):93.
- Qin XM, Qian F, Zeng DL, Liu XC, Li HM. 2011. Complete mitochondrial genome of the red-spotted tokay gecko (Gekko gecko, Reptilia: Gekkonidae): comparison of red-and black-spotted tokay geckos. Mitochondrial DNA. 22(5-6):176–177.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313.
- Starostová Z, Musilová Z. 2016. The complete mitochondrial genome of the Madagascar ground gecko Paroedura picta (Squamata: Gekkonidae). Mitochondrial DNA Part A. 27(6):4397–4398.
- Yan J, Tian C, Bauer AM, Zhou KY. 2016. The mitochondrial genome of the gold-dust day gecko, Phelsuma laticauda (Sauria, Gekkota, Gekkonidae). Mitochondrial DNA Part A. 27(1):73–75.
- Yan J, Tian C, Lv L, Bauer AM, Zhou KY. 2014. Complete mitochondrial genome of the San Lucan gecko, Phyllodactylus unctus (Sauria, Gekkota, Phyllodactylidae), in comparison with Tarentola mauritanica. Mitochondrial DNA Part A. 25(3):202–203.
- Yan J, Tian C, Zhou JL, Bauer AM, Lee Grismer L, Zhou KY. 2014. Complete mitochondrial genome of the Tioman Island rock gecko, Cnemaspis limi (Sauria, Gekkota, Gekkonidae). Mitochondrial DNA Part A. 25(3):181–182.
- Yan J, Zhou JL, Tian C, Zhou KY. 2009. Complete nucleotide sequence and gene organization of the mitochondrial genome of common house gecko, Hemidactylus frenatus. J Nanjing Normal Univ (Natural Science Edition). 32(4):77–82.
- Zhou KY, Li HD, Han D, Bauer AM, Feng JY. 2006. The complete mitochondrial genome of Gekko gecko (Reptilia: Gekkonidae) and support for the monophyly of Sauria including Amphisbaenia. Mol Phylogenet Evol. 40(3):887–892.
