Abstract
The complete mitochondrial genome of Japanese sponge crab Lauridromia dehaani was first determined and characterized from South China Sea. The L. dehaani mitogenome is 15,754 bp in length, and consists of 22 tRNA genes, two rRNA genes, 13 protein-coding genes (PCGs), and one control region. The nucleotide composition of L. dehaani mitogenome is significantly biased (A, G, T, and C was 36.23%, 9.71%, 35.12%, and 18.94%, respectively) with A + T contents of 71.35%. Two PCGs used an unusual initiation codon, and eleven PCGs were terminated with an abnormal stop codon. Three microsatellites were identified in L. dehaani mitogenome sequences. Phylogenetic tree showed that L. dehaani was first clustered with Dynomene pilumnoides.
Lauridromia dehaani, also known as Japanese sponge crab, is the most common dromiid species (McLay Citation2001). It always carries around a living piece of sponge on its back as a disguise so that it appears to be wearing a sponge hat. L. dehaani is known from a wide area of the Indo-West Pacific, and in the Asian area it is known from China, HongKong (Dai and Ng Citation1997), Taiwan, Japan and Philippines (McLay and Ng Citation2005). The sponge crab fashions its disguise by using its pincers to cut a piece of living sponge out of a larger sponge mass. It snips the sponge into a shape close to its own, and holds it up over its carapace using the rear legs. Recently new research unveils how to customize material into embodied cap and the individuality emerging in a making by it (Harada and Kagaya Citation2018). The results suggested that anything they can use to cover their bodies will do in a pinch, and interestingly, each crab had their own technique in styling their hat.
The specimens were collected from Huanqiu fishing port of Wenchang, China (N19°33′51.12″, E110°49′27.98″), and deposited in the marine crustacean specimen room (C20191207LD) in Qionghai research base of Hainan Academy of Ocean and Fisheries Sciences for reference, Muscle samples of L. dehaani were preserved in absolute ethanol for total DNA extraction.
The complete mitogenome of L. dehaani is 15,754 bp in length (GenBank Accession No. MT038417). The base content was 36.23% A, 9.71% G, 35.12% T, and 18.94% C. The 71.35% of (A + T) showed great preference to AT. It consists of 22 tRNA genes, two rRNA genes, 13 protein-coding genes (PCGs), and one control region (D-loop). Four PCGs (ND1, ND4, ND4L and ND5), eight tRNA genes and two rRNA genes were located on the light strand, the others were encoded by the heavy strand.
The 22 tRNA genes in L. dehaani mitogenome vary in length from 62 bp to 71 bp. tRNA-Leu and tRNA-Ser both have two type copies respectively. The 12S rRNA is 783 bp and located between tRNA-Val and D-loop, and the 16S rRNA is 1357 bp, located between tRNA-Val and tRNA-Leu. Except COX1 using an unusual CGA as the start codon, the others use a normal initiation codon ATN or TTG. Simultaneously, most PCGs were terminated with an unusual codon in addition to ATP8 and CYTB genes using a normal stop codon TGA. The control region is 566 bp, located between 12S rRNA and tRNA-Ile. Interestingly, we identified three microsatellites (SSRs) in L. dehaani mitogenome using MISA. Two (A)10 are located in ND1 genes, and a (TA)7 is sited in D-loop region.
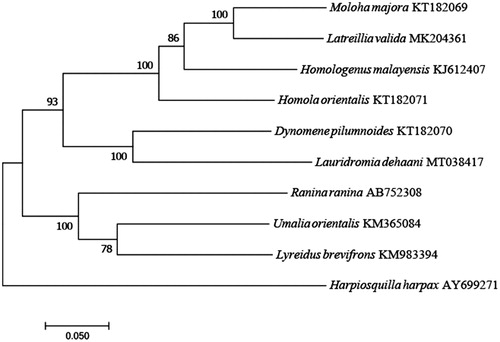
A maximum likelihood phylogenetic analysis with 8 crab mitogenomes placed L. dehaani among Podotremata species with 1000 bootstrap replicates. The result () showed that L. dehaani was first clustered with Dynomene pilumnoides, and further clarified the phylogenetic relationships of the family in Podotremata compared with the previous work (Bento et al. Citation2018).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bento MAG, Miranda I, Mantelatto FL, Zara FJ. 2018. Comparative spermatozoal ultrastructure and molecular analysis in dromiid crabs and their phylogenetic implications for Dromiidae and Podotremata (Decapoda: Brachyura). Arthropod Struct Dev. 47:627–642.
- Dai A-Y, Ng PK. 1997. Notes on an unusual specimen of Lauridromia dehaani (Rathbun, 1923) (Decapoda, Brachyura, Dromiidae) from Hong Kong. Crustac. 70(6):754–757.
- Harada K, Kagaya K. 2018. Individual behavioral type captured by a Bayesian model comparison of cap making by sponge crabs. Keita Harada, Naoki Hayashi, Katsushi Kagaya bioRxiv 330787; doi:10.1101/330787
- McLay C. 2001. A new genus and two new species of unusual dromiid crabs (Brachyura: Dromiidae) from Northern Australia. Rec Aust Mus. 53(1):1–8.
- McLay CL, Ng PK. 2005. On a collection of Dromiidae and Dynomenidae from the Philippines, with description of a new species of Hirsutodynomene McLay, 1999 (Crustacea: Decapoda: Brachyura). Zootaxa. 1029(1):1–30.