Abstract
The complete mitochondrial genome of Rhombodera longa Yang, 1997 was sequenced in this study (GenBank accession no. MT110155). This molecule is circular in shape with 15,886 bp length, containing 13 protein-coding genes (PCGs), 2 ribosomal RNA (rRNA) genes, 22 transfer RNA (tRNA) genes, and a non-coding control region. The AT content of the whole genome is 76.1% and the length of the control region is 636 bp with 78.2% AT content. This gene arrangement is consistent with the mitochondrial genomes of other praying mantises. Phylogenetic analysis revealed that R. longa clustered into a clade with Hierodula membranacea and found the genera Hierodula and Rhombodera are not monophyletic.
Mantodea are commonly known as predatory insects and includes more than 2500 extant species (Patel and Singh Citation2016). Species of the genus Rhombodera Burmeister possess a broad pronotum and are mainly distributed in the Oriental region (Schwarz and Roy Citation2019). Here, we determined the complete mitochondrial genome of Rhombodera longa Yang. The sampled specimen was collected from Jinghong in Yunnan, China (22°02′N, 100°55′E) in October 2018. It was identified based on the external morphology. The specimen was stored in the Collection of Insect Specimens of Shangluo University (voucher no. Ma-2018105). The complete mitochondrial DNA sequence was determined by Illumina HiSeq 2500 Sequencing System (Illumina, San Diego, CA, USA). In total, 5.5 G raw reads were obtained, quality-trimmed, and assembled using MITObim version 1.7 (https://github.com/chrishah/MITObim) (Hahn et al. Citation2013).
The complete mitochondrial genome of R. longa was 15,886 bp in total length. It is deposited in the GenBank database with an accession number MT110155. The overall base composition was 40.1% A, 36.0% T, 14.5% C, 9.4% G, with an A + T ratio of 76.1%. The full mitochondrial genome contains 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), 2 ribosomal RNAs (rRNAs), and a putative control region. The gene arrangement of R. longa is similar to that normally observed in Mantidae, the family of praying mantises (Ye et al. Citation2016; Zhang and Ye Citation2017). Most PCGs in R. longa have the conventional start codon for invertebrate mitochondrial PCGs (ATN), with the exception of cox1 (TTG). Most of the PCGs terminate with the stop codon TAA, whereas cox2 and nd5 end with the incomplete codon T––. Twenty-four genes are encoded on the major strand (J-strand), whereas all others are encoded on the minor strand (N-strand). Nine PCGs are encoded on the J-strand and four (nd5, nd4, nd4L and nd1) on the N-strand. Two rRNA genes (rrnL and rrnS) are located at the trnL1/trnV and trnV/control regions respectively, and both rRNA genes are encoded on the N-strand. The lengths of rrnL and rrnS are about 1310 and 747 bp, with A + T content of 79.0% and 75.9% respectively. The length of the control region is 902 bp, with the AT content of this region up to 78.2%.
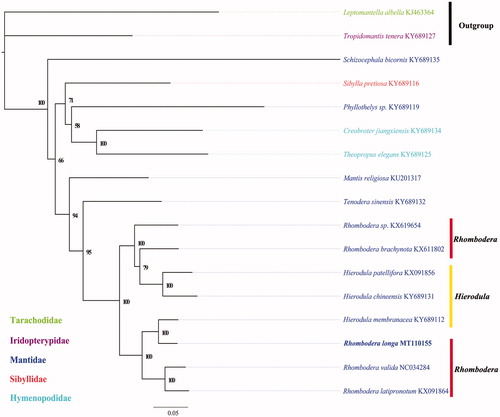
To validate the phylogenetic position of R. longa, 13 mitochondrial protein-coding gene sequences were extracted from 17 closely related taxa of Mantodea. A phylogenetic tree was constructed using the maximum-likelihood method through IQtree 1.6.8 (Nguyen et al. Citation2015). Results show that R. longa clusters into a clade with Hierodula membranacea, and then clustered into a clade with R. valida + R. latipronotum (). The monophyly of the genera Hierodula and Rhombodera was not supported, which is consistent with previous studies (Zhang et al. Citation2018; Jia et al. Citation2019). This indicates that further taxonomic study of closely related genera Hierodula and Rhombodera is required. In conclusion, we obtained and described the complete mitochondrial genome of R. longa, which constitutes a valuable resource for population genetic study and identification efforts on this species.
Acknowledgements
We are grateful to Prof. J. R. Schrock (Emporia State University, USA) for revising this manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129–e129.
- Jia Y-Y, Zhang L-P, Xu X-D, Dai X-Y, Yu D-N, Storey KB, Zhang J-Y. 2019. The complete mitochondrial genome of Mantis religiosa (Mantodea: Mantidae) from Canada and its phylogeny. Mitochondrial DNA Part B. 4(2):3797–3799.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274.
- Patel S, Singh R. 2016. Updated checklist and distribution of Mantidae (Mantodea: Insecta) of the world. Int J Res Stud Zool. 2(4):17–54.
- Schwarz CJ, Roy R. 2019. The systematics of Mantodea revisited: an updated classification incorporating multiple data sources (Insecta: Dictyoptera). Ann Soc Entomole France (NS). 55(2):101–196.
- Ye F, Lan XE, Zhu WB, You P. 2016. Mitochondrial genomes of praying mantises (Dictyoptera, Mantodea): rearrangement, duplication, and reassignment of tRNA genes. Sci Rep. 6:25634.
- Zhang HL, Ye F. 2017. Comparative mitogenomic analyses of praying mantises (Dictyoptera, Mantodea): origin and evolution of unusual intergenic gaps. Int J Biol Sci. 13(3):367–382.
- Zhang LP, Yu DN, Storey KB, Cheng HY, Zhang JY. 2018. Higher tRNA gene duplication in mitogenomes of praying mantises (Dictyoptera, Mantodea) and the phylogeny within Mantodea. Int J Biol Macromol. 111:787–795.