Abstract
The complete mitochondrial genome of the hybrid of O. stewartii (female) and S. younghusbandi (male) was sequenced and characterized in this study. The complete hybrid mitochondrial genome is 16,649 bp in length, including 2 rRNAs, 13 protein-coding genes, 22 tRNAs and 2 non-coding regions (OL and D-loop region). The results discovered that 99.89% sequence identity between the hybrid and its female parent, which confirmed the maternal inheritance pattern followed by the mitochondrial genome of the hybrid. The phylogenetic tree showed that the hybrid was relatively closer to O. stewartii. Our data would provide a valuable resource for further studies on mitochondrial inheritance mechanism of Schizothoracinae fishes and provide new insights for better conservation plans for fishery resources.
Hybridization is a quite common phenomenon in nature, which playing a significant role in evolution in both plants and animals (Michael L. Arnold Citation1997). Hybridization has also been observed in Schizothoracinae fishes in recent years (Ma et al. Citation2018; Zhang et al. Citation2019). Oxygymnocypris stewartii (Lloyd.1908) and Schizopygopsis younghusbandi (Regan.1905) are belong to the subfamily of the Schizothoracinae. These species are Widely distributed in the Lhasa River with an average altitude over 3800 m (Chen and Cao Citation2000). Recently, due to the damming of rivers and overfishing, the natural resources of these species have dropped significantly, and O.stewartii has been assessed as endangered fish in Red List of China’s Vertebrates (Jiang et al. Citation2016). In this study the complete mitogenome of the hybrid of O. stewartii (female) and S. younghusbandi (male) was sequenced for the first time.
The specimens were obtained from the Lhasa River (N29°40′27.00″, E91°23′28.53″) at an altitude of 3650 m in 2019 and stored in 95% ethanol with accession number: 20190608JK01. The total genomic DNA was extracted from the pelvic fin with a traditional phenol-chloroform method (Sambrook and Russell Citation2001). The mitogenome was amplified by 19 pairs primers (Zhang et al. Citation2016) and annotated by DOGMA (Wyman et al. Citation2004). Also, the mitochondrial genome sequence of the hybrid loach was aligned by BLAST. The neighbor-joining (NJ) tree of the hybrid was constructed using Mega7.0 (Kumar et al. Citation2016).
The entire mitogenome sequence of the hybrid loach was 16,649 bp in length (GenBank number MT130395), including 2 ribosomal RNA (rRNA), 13 protein-coding genes, 22 transfer RNA (tRNA) genes and 2 control regions. The arrangement of encoded genes was similar to the other Schizothoracinae fishes (Qiao et al. Citation2013; Ji et al. Citation2015; Zhang C et al. Citation2019). The overall base composition of the heavy strain was A (30.8%), T (30.42%), C (19.3%), G (19.49%), and the content of A + T in the complete genome was 61.22%. In addition, 12 PCGs, 14 tRNA genes and two rRNA genes were located on the heavy strand (H-strand), while one PCG (ND6) and eight tRNA genes were encoded on the light strand. The sequence alignment of mitochondrial genomes between the hybrid and its female parent revealed a total of 16 variable sites, especially 9 base mutations sites are located in ND1, COX2, ATP6, ND5 and CYTB.
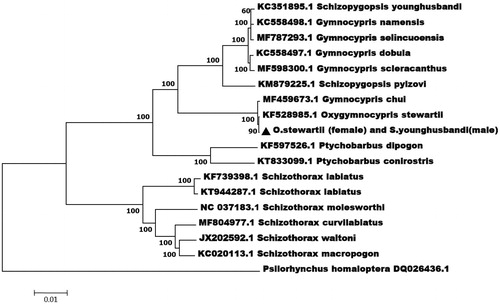
To further explore the taxonomic status of the hybrid loach, we performed the phylogenetic relationship of the hybrid loach stock with other Tibetan highland loach fishes using the complete mitochondrial genome sequence. The phylogenetic tree () was constructed with NJ method using 13 species of fish. The results showed that the hybrid was relatively closer to O. stewartia (female parent) than the other species, which is consistent with the matrilineal inheritance mechanism. This is the first report on mitochondrial genome of the hybrid of O. stewartii (female) and S. younghusbandi (male), which provides data for studies on adaptive evolution and genetic diversity in Schizothoracinae fishes.
Figure 1. The consensus phylogenetic relationship of the hybrid species of O. stewartii (female) and S. younghusbandi (male) with other fishes. Phylogenetic tree based on the complete mitochondrial genome sequences was constructed by neighbour-joining method. GenBank accession numbers of mitogenomic sequences for each taxon are shown in parentheses.
Acknowledgements
The authors would like to thank Jiawen Xu for the assistance in sample collection and identification.
Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Additional information
Funding
References
- Michael L. Arnold. 1997. Natural hybridization and evolution. Oxford University Press. 1–232.
- Chen Y, Cao W. 2000. Schizothoracinae. In: Fauna Sinica Osteicthtyes Cypriniformes III. Beijing: Science Press. 1–338.
- Ji W, Zhang GR, Wei KJ, Guo SS, Guo XZ, Wei QW. 2015. Characterization of the complete mitochondrial genome of Oxygymnocypris stewartii (Cypriniformes: Cyprinidae). Mitochondrial DNA. 26(5):684–685.
- Jiang Z, Jianping J, Wang YZ, Zhang E, Zhang YY, Xie F, Cai B, Cao L, Zheng GM, Dong L, et al. 2016. Red list of China’s vertebrates. Biodiver Sci. 24(5):500–551.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Ma B, Li L, Wang J, Gong J, Zhang C, Ji F, Li B. 2018. Morphological and COI gene barcode analyses of Schizothorax waltoni, S. o’ connori and their natural hybrids in the Yarlung Zangbo River. J Fishery Sci China. 25(4):753.
- Qiao H, Cheng Q, Chen Y, Ren G. 2013. The complete mitochondrial genome sequence of Schizopygopsis younghusbandi (Cypriniformes: Cyprinidae). Mitochondrial DNA. 24(4):388–390.
- Sambrook JF, Russell DW. 2001. A laboratory manual. 3rd ed. New York (NY): Cold Spring Harbor Laboratory.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 20(17):3252–3255.
- Zhang J, Chen Z, Zhou C, Kong X. 2016. Molecular phylogeny of the subfamily Schizothoracinae (Teleostei: Cypriniformes: Cyprinidae) inferred from complete mitochondrial genomes. Biochem Syst Ecol. 64:6–13.
- Zhang C, Zhou J, Liu F, Mou Z, Chen M. 2019. The complete mitochondrial genome of the hybrid of Schizothorax oconnori (♀) × Schizothorax waltoni (♂). Mitochondrial DNA B. 4(1):1983–1985.
