Abstract
Cirrhinus mrigala is a fish introduced from India to the south China in 1980s, and a large number of this species is found in the Three Gorges Reservoir of the upper Yangtze River now, which might pose a great threat to the indigenous fish. In this study, we sequenced and characterized the complete mitochondrial genome of C. mrigala from the Three Gorges Reservoir. The complete mitochondrial genome was 16,595 bp in length and contained 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA) and one non-coding control region (D-loop). The overall nucleotide composition was 32.05% (A), 24.48% (T), 15.49% (G) and 27.98% (C). The phylogenetic tree indicated that C. mrigala was related to Sinilabeo rendahli, Abbottina rivularis and Pseudogyrinocheilus prochilus. The results can provide basic genetic information for subsequent population genetic and phylogenetic researches, which will be helpful for understanding the genetic resource dynamic of C. mrigala in areas outside of its native distribution.
Cirrhinus mrigala (genus: Cirrhinus, subfamily: Labeoninae, family: Cyprinidae) is a native fish of the Indus and Ganges river systems (Misra Citation1962; Chauhan et al. Citation2007). It was introduced successfully from India to China in 1980s, and widely cultivated with other fishes under polyculture system in the south China now (Chen et al. Citation2004). The growth rate of C. mrigala is fast, whose fry can grow to more than 400 g in the first year, and the largest individual can reach more than 3000 grams (Chondar Citation1999). C. mrigala is an omnivorous fish with a wide range of food source, mainly feeding on plant debris (Yashpal et al. Citation2009). C. mrigala’s fertility is high, which increases with age, usually 100,000–150,000 eggs per kilogram of body weight (Chondar Citation1999). This species is resistant to hypoxia and has strong disease resistance, it can grow normally in reservoirs, lakes, ponds and rivers (Chen et al. Citation2004). Therefore, a large number of C. mrigala is found in the Three Gorges Reservoir of the upper Yangtze River may pose a great threat to the indigenous fish. In this study, we sequenced and characterized the complete mitochondrial genome of C. mrigala from the Three Gorges Reservoir, it can provide basic genetic information for subsequent population genetic and phylogenetic researches, which will be helpful for understanding the genetic resource dynamic of C. mrigala in areas outside of its native distribution.
Sample of C. mrigala was collected from Taipingxi (N30°52′28″ E110°57′44″) section of the Three Gorges Reservoir in 2019. The collected sample was preserved in 95% alcohol and deposited at −20 °C in Specimen Room of School of Life Sciences, Jianghan University (Sample number: 20190812001). Total DNA was extracted from dorsal muscle using the phenol-chloroform method (Sambrook and Russell Citation2001). The mitochondrial genome was sequenced by the next-generation sequencing technology. We de novo assembled complete mitochondrial genome by NOVOPlasty ver 2.6 (Dierckxsens et al. Citation2017). Protein-coding genes and rRNA genes were created by web server DOGMA (Wyman et al. Citation2004), and tRNA genes was predicted by using the program ARWEN (Laslett and Canback Citation2008).
The complete mitochondrial genome of C. mrigala was 16,595 bp in length (GenBank accession number: MT136763) and contained 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S rRNA and 16S rRNA), 22 transfer RNA genes (tRNA) and one non-coding control region (D-loop). The overall nucleotide composition was 32.05% (A), 24.48% (T), 15.49% (G) and 27.98% (C), and the A + T content (56.5%) was much higher than G + C content (43.5%). Except for the ND6 gene and eight tRNA genes (tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UGA), tRNAGlu and tRNAPro), others were encoded on the heavy strand. Twelve of thirteen protein-coding genes were initiated with a typical ATG codon, except COI gene was started with GTG. The stop codon TAA was found in six genes (COX1, COX3, ATP6, ND4L, ND5 ND6), and TAG was found in three genes (ND1, ATP8, ND3), while the remaining four genes (COX2, ND2, ND4, CYTB) terminated with a single base T. The total length of coding sequence was 11,413 bp, which encoded 3794 amino acids.
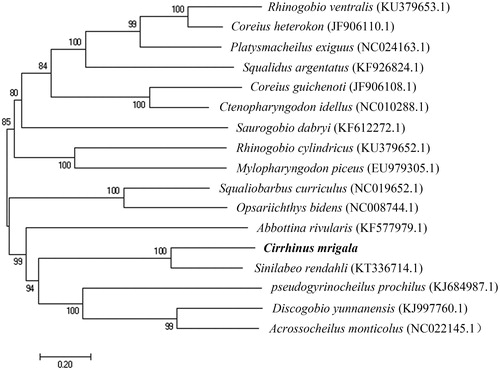
In order to investigate the phylogenetic relationships between C. mrigala and other 16 fish species that might have niche competition with C. mrigala in the Three Gorges Reservoir, a phylogenetic tree was constructed using Neighbor-Joining method by MEGA7 (Kumar et al. Citation2016) based on the whole mitogenome sequences. All the data were downloaded from NCBI GenBank, and the accession numbers were shown on the . The results indicated that C. mrigala was related to Sinilabeo rendahli, Abbottina rivularis and Pseudogyrinocheilus prochilus.
Disclosure statement
There are no conflicts of interest for all the authors including the implementation of research experiments and writing this article.
Additional information
Funding
References
- Chauhan T, Lal KK, Mohindra V, Singh RK, Punia P, Gopalakrishnan A, Sharma PC, Lakra WS. 2007. Evaluating genetic differentiation in wild populations of the Indian major carp, Cirrhinus mrigala (Hamilton–Buchanan, 1882): Evidence from allozyme and microsatellite markers. Aquaculture. 269(1–4):135–149.
- Chen K, Xiao D, Wen Z, Lin S, Kuang G. 2004. A comparative study on the biological characteristics of Cirrhinus mrigala and Labco rohita. Inland Fisheries. (6):37–38. (In Chinese)
- Chondar SL. 1999. Biology of finfish and shellfish. 1st ed. West Bengal: SCSC Publishers (India); p. 52–65.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874.
- Laslett D, Canback B. 2008. ARWEN, a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175.
- Misra KS. 1962. An aid to the identification of the common commercial fishes of India and Pakistan. Rec Indian Mus. 57:1–320.
- Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual. 3rd ed. New York (NY): Cold Spring Harbor Laboratory Press.
- Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with dogma. Bioinformatics. 20(17):3252–3255.
- Yashpal M, Kumari U, Mittal S, Mittal AK. 2009. Morphological specializations of the buccal cavity in relation to the food and feeding habit of a carp Cirrhinus mrigala: a scanning electron microscopic investigation. J Morphol. 270(6):714–728.